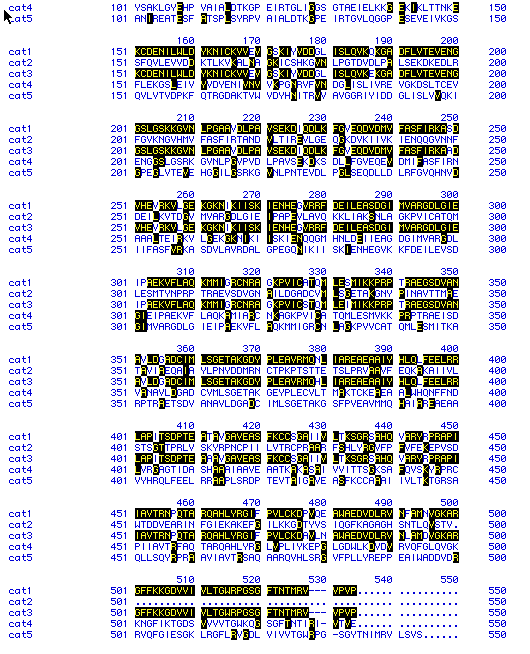MacDNAsis of My Favorite Protein: Pyruvate
Kinase
This webpage was created as an assignment for an undergraduate course
at Davidson College in Molecular Biology.
Click here to find out more about this protein
and the sequence information from the Genbank Search. After finding
out the cDNA sequence of the Homo sapiens Pyruvate Kinase gene,
I used MacDNAsis to translate the sequence and determine its Open Reading
Frame, molecular weight, hydrophobicity, predicted secondary structure,
and the extent of its relatedness to the other four species' protein structures.
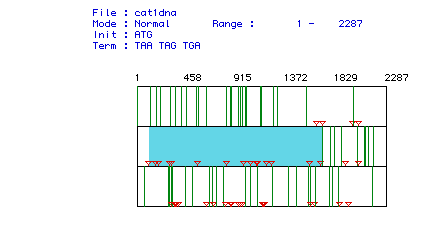 Figure 1: Area highlighted in blue corresponds to the largest Open
Reading Frame of Human Pyruvate Kinase cDNA. Each arrow represents
a possible start/stop coding sequence. Through process of elimination
the longest interval between the green lines was selected and is the ORF
corresponding to the protein.
Figure 1: Area highlighted in blue corresponds to the largest Open
Reading Frame of Human Pyruvate Kinase cDNA. Each arrow represents
a possible start/stop coding sequence. Through process of elimination
the longest interval between the green lines was selected and is the ORF
corresponding to the protein.
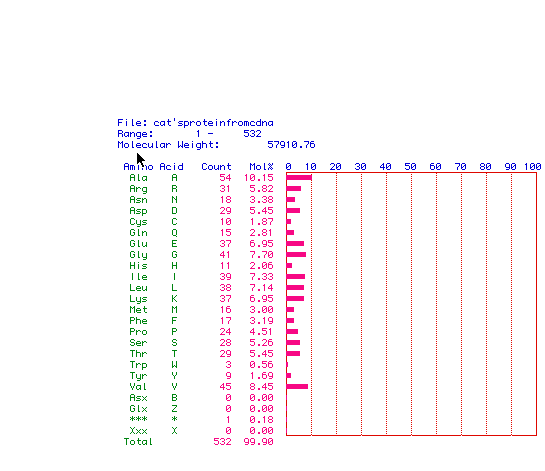
Figure 2: Analysis of Amino Acid content of Human Pyruvate Kinase.
The molecular weight of this protein is 57.9 Kbp. The break down
of how many of each amino acid contained in the protein is listed on the
left with a bar graph representation on the right.
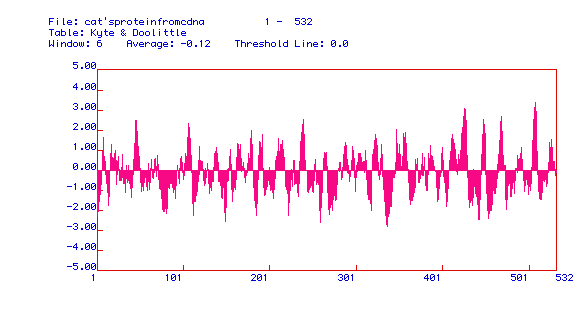
Figure 3: Kyte and Doolittle Hydropathy plot of Human Pyruvate
Kinase. This particular figure is very useful in discovering if a
particular protein has integral membrane possibilities. Integral
membrane proteins must have hydrophobic domains which span the hydrophobic
interior of the membrane. Area above 0.00 possibly corresponds to
hydrophobicity when the peak reaches the 2.00 range. This particular
hydropathy plot shows that pyruvate kinase has many possible integral membrane
domains and could be an integral membrane protein. This is consistent
with Pyruvate kinase's role in cellular respiration.
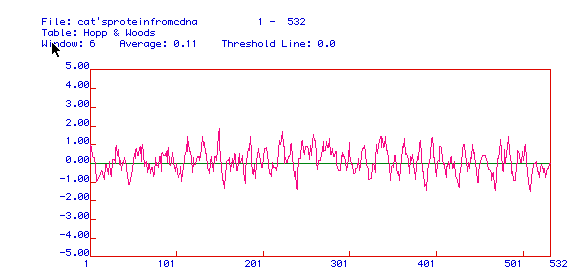
Figure 4: Hopp and Woods Hydropathy plot of Human Pyruvate Kinase.

Figure 5: Chou, Fasman and Rose prediction of the secondary protein
structure of Human Pyruvate Kinase. The key decoding the colors are
listed above the figure. Blue corresponds to an alpha helix, red
and white stripes corresponds to a beta sheet, green corresponds to a turn,
and black/white stars correspond to coils.
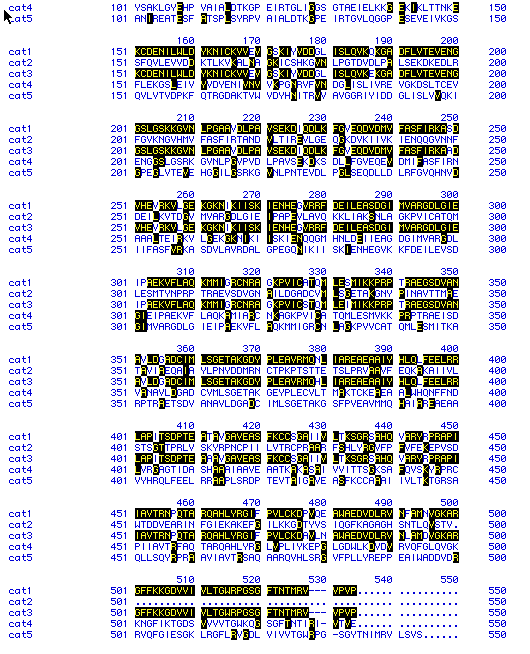
Figure 6: Sequence alignment of five different species' Pyruvate
Kinase proteins. Black boxes indicate similarity between sequences.
Dashed lines are included as spacers to increase similarities between the
proteins.
key: cat1 = Homo sapiens (click here to see
DNA sequence)
cat2 = Saccharomyces cerevisiae
(click here to see DNA sequence)
cat3 = Mus musculus (click
here to see DNA sequence)
cat4 = Drosophila melanogaster
(click here to see DNA sequence)
cat5 = Rattus norvegicus (click
here to see DNA sequence)
Finally, using the MacDNAsis program, I generated a phylogenetic tree
illustrating the evolutionary relationship between the five proteins.
As would be expected cat1 (human) and cat3 (mouse) are the most closely
related as they are both mammals. The similarity between the proteins
dropped off quickly between human, mouse and rat and the fruitfly and yeast
were by far the least related.
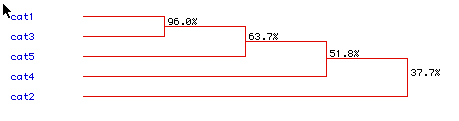
Figure 7: Phylogenetic Tree comparing cat1 (Homo sapiens), cat2 (Saccharomyces
cerevisiae), cat3 (Mus musculus), cat4 (Drosophila melanogaster), and cat5
(Rattus norvegicus) protein.
RETURN TO MY MOLECULAR
HOME PAGE
CLICK HERE
Return to the Molecular
Biology Home Page, or Immunology
Home Page.
Return
to Davidson College Biology Department Home Page
Return
To Biology Course Materials

Comments? Questions? Email me cakizer@davidson.edu
© Copyright 2000 Department
of Biology, Davidson College, Davidson, NC 28036.