This page is part of an
undergraduate assignment at Davidson College
My Favorite Yeast Proteins: Understanding the Function of
YEL068C and MATA1.
††††††††††††††††††††††††††††††††††††††††††
The
mysteries of a gene are not elucidated when only information about genesí nucleotide
sequence has been obtained. The sequence can give clues to a genesí function,
but it cannot reveal the full story. The protein products of gene can be post
transcriptionally modified, which can change its function. Secrets of a
proteins function are better understood by examining its amino acid sequence
and by understanding the roles of the proteins that it with which it interacts.
This final web project reports the results of database searches about the
protein coded by the genes YEL068C and MATA1, and it includes a hypothetical
experiment design that could be used to better understand the function of these
proteins.
Stan
Fields, Benno Schwikowski & Peter Uetz 2000: PDF files
Stan Fields, Benno Schwikowski, and Peter Uetz
compiled 2709 interactions among 2039 yeast proteins to generate an integrated
protein circuit of 1, 548 proteins.†
YEL068C: http://bio.davidson.edu/courses/genomics/Benno/NB_Figure1color.pdf
This
circuit is color coded by lines and boxes surrounding the proteins. The lines
represent: red= cellular role and subcellular
localization of interacting proteins are identical; blue=localizations are identical but function differ; green=cellular roles are identical but localizations
differ; black=cellular role and localization are different or
unknown; gray= not explained in paper. The
boxes represent: blue= membrane fusion; gray=chromatin structure; green=cell structure; yellow=lipid
metabolism; red=cytokinesis; butter color= not explained in paper.
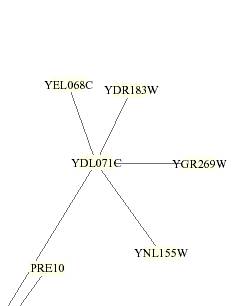
YEL068C was located on a remote branch of the circuit
but clusters with the proteins coded by DR183W, YDL071C, YGR269W, PRE10, and
YNL155W. By using the SGD Full Search search engine, I was able to
determine the role of the genes with which YEL068C clusters with in hopes of
being able to make an inference about YEL068Cís role. YEL068C only directly
interacts with YDL071C, but this gene is unannotated. The line indicated the
interaction is gray, which is not explained in the table, so no information is
revealed about the nature of the interaction. DR18W, YGR269W, and YNL115W are
also unannotated. The SGD revealed that PRE10 is a multipcatalyitc endopepetidase
involved in ubiqutin-dependent protein degradation (Hochstrasser 1996).† Ubiquitin modification is related to cell
cycle, class I antigene processing, signal transduction pathways, receptor
mediated endocyctosis, and 26S proteasome degradation of bounded proteins
(Hochstrasser 1996). †Both YEL068C and PRE10
are connected through a distant circuit involving GNA1
(not pictured). Further investigation could explain YEL068C role by
understanding the role of PRE10. †YEL068C was in a butter colored box, which has no explanation
associated with it, and thus no information on the protein could be derived
through this option.
†
MATA1: http://bio.davidson.edu/courses/genomics/Benno/NB_Figure1color.pdf
This
circuit is color coded by lines and boxes surrounding the proteins. The lines
represent: red= cellular role and subcellular
localization of interacting proteins are identical; blue=localizations are identical but function differ; green=cellular roles are identical but localizations
differ; black=cellular role and localization are different or
unknown; gray= not explained in paper. The
boxes represent: blue= membrane fusion; gray=chromatin structure; green=cell structure; yellow=lipid
metabolism; red=cytokinesis; butter color= not explained in paper.
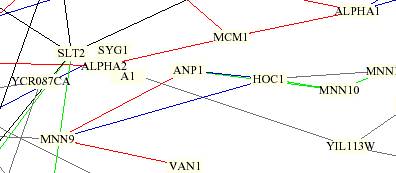
A1 is in a butter-colored box and is connected to YIL113W
by a gray line. Although it is difficult to determine from the picture, I
believe that it may be connected to ALPHA1 too. By completing a SGD Full Search I
was able characterize the function of some of the genes clustered around MATA1.
†MATA2, ALPHA1, and
MCM1
are all involved in the mating process. This result supports the
research on MATA1, which is included on my MFYG webpage that states that MATA1
is a translation factor that facilitates the switch of yeast mating type. YIL113
is unannotated (SGD 2000). It would be interesting to determine
if this gene is involved in the mating type switch. †SLT2
is a MAP kinase protein involved in protein amino acid
phosphorylation and signal transduction pathways in the nucleus (Lee 1993). It
is possible that this gene may be involved in signaling to insight the mating
type switch, but that assumption is unclear. ††ANP1
is a protein amino acid glycosylation, which is located in the
endoplasmic reticulum and aids in retention of glycosyltransferases in the
Golgi as well as osmotic sensitivity (SGD 2000).†† I am not sure how this could relate to the mating type switch.
I also searched the several pdf files that are based
on experiments surrounding the processes of aging, degradation, and membrane formation
for the genes YEL068C and MATA1. †YEL068C is on circuit for the aging, degradation, and membrane formation
experiments. †It clustered and
interacted with the same proteins for all their targeted circuits.
Aging circuit:
http://bio.davidson.edu/courses/genomics/Benno/aging.pdf
Membrane circuit: http://bio.davidson.edu/courses/genomics/Benno/membrane.pdf
Degradation circuit: http://bio.davidson.edu/courses/genomics/Benno/degradation.pdf
YEL068C
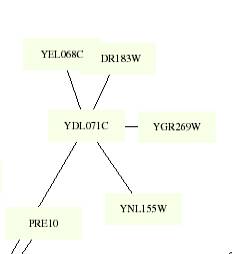 Th
Th
This figure is color coded by the following: Aging: Gray, Pol-II-transcription:
LightBlue, Protein-translocation: Yellow, Protein-synthesis: OrangeRed, Other: EmeraldGreen,
Phosphate-metabolism: Black, Vesicular-transport:
IndianRed,
Cytokinesis: LightGray
YEL068C clustered with the same group of genes in each
of the targeted circuits and was located in the same remote location. The color
box surrounding provides no additional information about the protein. Thus, I
do not believe that it has much to do with the aging, degradation, or membrane
formation processes.
MATA1
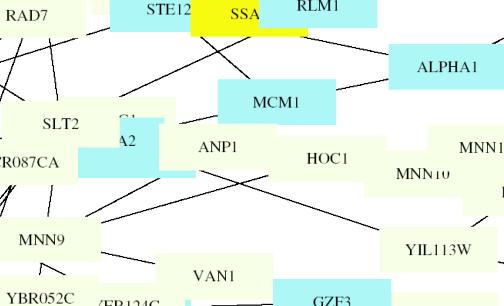
This figure is color coded by the following: Aging: Gray, Pol-II-transcription:
LightBlue, Protein-translocation: Yellow, Protein-synthesis: OrangeRed, Other: EmeraldGreen,
Phosphate-metabolism: Black, Vesicular-transport:
IndianRed,
Cytokinesis: LightGray
MATA1 is located directly behind A2 (MATA2). Thus I am
unable to determine the color of the box surrounding it and cannot use the
color to make inferences about the protein function. I also was unable to
determine which genes it directly interacted with as denoted by a line. I can
tell that it is located near the center of the circuit and clusters with A2,
ANP1, MCM1, YIL113, ALPHA1, SLT2.†
Yeast Two Hybrid Database
The Yeast Two Hybrid Database was developed by Uetz
et.al. MATA1 is not listed on this database, but YEL068C is a reported bait
protein.
YEL068C (prey) interacts with YDL071C (bait).† YDL071C is yet to be annotated (YPD 2000)
so it is impossible to make any assumptions about the function of YEL068C based
on this interaction.
The Triples Database is a database of TRansposon-Insertion
Phenotypes, Locationization,
and Expression in Saccharomyces.
By utilizing a transposing based approach, researchers are able to develop
three types of function data: 1) when a gene is expressed during the cell
cycle, 2) disruption phenotypes, and 3)†
subcellular localization.
YEL068C and MATA1 are not in Triples Database.
Database
for Interacting Proteins (DIP)
This database reveals protein-protein interaction for
a queried gene. My search on DIP yielded no results for YEL068C and MATA1.
Enzymes and
Metabolic Pathways Database (EMP)
This database allows you to enter a substrate and find
out what metabolic pathway it is involved in. My search for YEL068C and MATA1
yield no results on EMP database.
WIT
Database
WIT reveals protein function and the metabolic pathway
it is involved with for multiple organisms. WIT did not produce any results for
YEL068C and MATA1.
This database consists of data derived from protein chips of
kinases. YEL068C and MATA1 are not included. It is not surprising MATA1 is not
included because it is not a kinase. Just because YEL068C is not included on
this database does not allow the assumption that YEL068C is not a kinase
because YEL068C may not have been included on the protein chip.
Conclusions
The results produced for MATA1 from the Fields, Schwikowski,
and Uetz PDF files are consistent with the expected function as a protein
involved in the mating type switch. However, I was disappointed to discover
that MATA1 was not included on many of the other databases.
The results produced for YEL068C by the Fields, Schwikowski,
and Uetz PDF files and the Yeast Two Hybrids really revealed little about the function
of the protein mainly because the only protein YEL068C interacts with is also unannotated.
Hypothetical Experiment to Better
Understand the Protein Function of YEL068C
In order to understand the function of YEL068C, I
would like to crystallize the protein. This process would be very difficult.
First 1 milligram of YEL068C would need to be purified. To purify such a large
quantity of †the protein you would need
to overproduce it, perhaps by cloning in an extra copy and growing large
cultures. Then you would need to lyse the cells and purify. Then, I would use
nuclear magnetic resonance (NMR) to determine the location of the atoms. From
this model, I would look for unique folding patterns and molecules. Perhaps the
conformation and unique folding patterns and/or molecules could suggest possible
targets for the protein or it would resemble a class of proteins with a unique
function.