This web page was produced as an assignment for an undergraduate course at Davidson college.
The Gene for Dyslexia is revealed
News Report - "Researchers link gene to Dyslexia"
On Tuesday, August 26, 2003, ABC (Austrialian Broadcasting Corporation) news online published an article stating that Finnish researchers had found a gene that is important in causing dyslexia. Dyslexia, they stated, is the most common learning disorder among children and affects 3 to 10% of the population in the United States. Dyslexia is characterized by problems with word recognition and reading. As ABC reports, evidence has shown that people with dyslexia use the right side of their brain for word processing rather than the left side of the brain, as people without dyslexia instinctively do. The Finnish investigators began investigations with a family who had several dyslexic members. The father had severe learning disabilities and his three children have disabilities as well. As a comparison, the team used 20 unreleated families in their study. The scientists found that the first family had a gene "DYXC1" that was disrupted in those members with dyslexia. In addition, another unrelated dyslexic family had a truncated version of the same gene. ABC reports that based on this data, the researches conclude that DYXC1 is a strong candidate gene for dyslexia, especially given the evidence that dyslexia is genetic.
This article, published ahead of the e-publication of the reviewed article, gave a good summary of the information provided. Although the evidence was slightly exaggerated and not much emphasis was place on the fact that this gene is located in one of six regions of the genome linked to dyslexia, ABC provided sufficient information to explain to the public that progress is being made in the field of dyslexic genetics. They even included facts such as, "The researchers said that this gene is unlikely to explain all cases of dyslexia."
For more information on dyslexia see the International Dyslexia Foundation site
PNAS article (Taipale et al 2003)
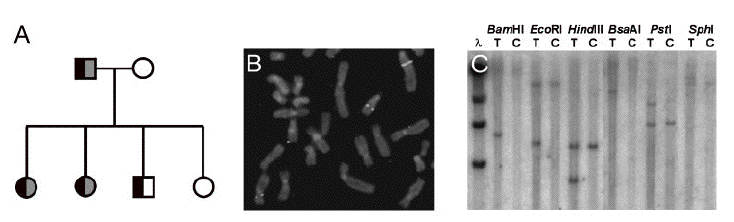
Taipale et al published "A candidate gene for developmental dyslexia encodes a nuclear tetratricopeptide repeat domain protein dynamically regulated in brain" stating that the group had discovered a "candidate susceptiblility gene for developmental dyslexia." Using the family described above in the ABC article, the researchers found a BAC containing the translocation region in the family (see Figure 1). With only one unknown gene in that particular BAC, the researchers dubbed the gene DYX1C1. A mouse homolog was found which was 70% identical to the human protein. The researchers used three different methods to support their idea that the DYX1C1 gene is related to dyslexia. First, the researchers showed that the gene was disrupted by translocation in the family that was plagued with dyslexia. In addition, they showed that two SNPs (or single nucleotide polymorphism) significantly associated independently with the dyslexia phenotype using a TDT (transmission disequilibrium test). Lastly, the researchers showed that a third SNP located at the transcription factor binding sequence for Elk-1, HSTF, and TFII-I. Elk-1 is a transcriptional activator expressed in rat brain neurons and it's activation has been linked to learning in rats. Expression of DYX1C1 was abundant in human brain, lung, kidney, and testis. In addition, DYX1C1 was shown to localize to the nucleus through imunoreactivity in human brain tissue and immunoflourescence in monkey kidney COS-1 cells.

Figure 1. (A) The pedigree for the family affected by dyslexia. The father has severe dyslexia, and three of the children are thought to be affected. The boy cannot be confidently classified as dyslexic since he has shown some more serious learning disorders. (B) FISH using BAC (bacterial artifical chromosome) clone 178D12, showing hybridization on chromosome 15, der(15) and der(2). (C) Southern blot hybridization using the probe from BAC 178D12 shows translocational rearrangements. Patient T is the translocation subject, while patient C is the control subject. Six restriction enzymes were used to show the translocation. Figure reproduced from Taipale et al, 2003. Permission requested.
Given this information, the claim of the author's seems valid, that Dyx1c1 should be considered as a candidate gene for dyslexia susceptibility. This study is very interesting because it is the first study that has found a gene directly related to dyslexia. Although several regions of chromosomes have been linked to dyslexia inheritance, this study is the first to provide a gene to support the inheritance theory. In fact, this gene is not located in the region of chromosome 15 that has been shown to be linked to dyslexia inheritance. DXY1C1 is located about seven megabases from the region of chromosome 15, DYX1, suspected to be involved in dyslexia. Given this information, DYX1C1 my correspond to DYX1 or it may be another gene that whose linkage to dyslexia had not been noticed until this paper. The functionality of DYX1C1 is unclear. The amino acid sequence doesn't provide much information regarding the cellular role of DYX1C1. The protein contains three TPR (tetratricopeptide repeat) motifs, but they are found in a wide variety of proteins in various phyla. However, it is interesting that the protein differs more in humans from gorillas and orangutan than from chimpanzee and pygmy chimpanzee proteins, which may imply important evolutionary differences in brain development between the primates. Based on the human protein variation, however, the researchers conclude that the Elk-1 binding site and possible the C-terminal of the protein are the most functionally important. Based on their data, the researchers hypothesize that DYX1C1 is involved "dynamically in the functional cell state, changing in the face of metabolic challenge." Overall, DYX1C1 is just the first gene found to be related to dyslexia. Based on the frequency of dyslexia in the population and the number of loci already linked to dyslexia inheritance, we can conclude that several more genes function in the process of creating dyslexia and would lead us to believe that there is a wider variety of phenotypes for dyslexia than one would initally think.
References
ABC NewsOnline. Researchers link gene to dyslexia. August 26, 2003. <www.abc.net.au/news/newsitems/s931922.htm>
Taipale, M, N Kaminen, J Nopola-Hemmi, T Haltia, B Myllyluoma, H Lyytinen, K Muller, M Kaaranen, P Lindsberg, K Hannula-Jouppi, and J Kere. A candidate gene for developmental dyslexia encodes a nuclear etratricopepetide repeat domain protein dynamically regulated in brain. Proc Natl Acad Sci U S A. 2003 Sep 3 [Epub ahead of print] <www.pnas.org/cgi/reprint/1833911100>
Questions or comments? E-mail Sarah Baxley