This webpage was produced as an assignment for an undergraduate course at Davidson College
My Favorite Yeast Expression
Annotated Gene: ERG25
Background: My known gene ERG25 is an essential component in the ergosterol biosynthesis pathway. The ERG25 protein is a membrane bound protein called C-4 methyl sterol oxidase that is localized in the endoplasmic reticulum and plasma membrane. The protein is involved in the formation of sterols--derivatives of steroids--from simpler compounds.
Mining microarray databases can reveal more information about the function of this gene. Investigating the Expression Connection database revealed several conditions under which ERG25 produced dramatic levels of induction or repression compared to normal conditions. Of all the expression conditions available to mine at Expression Connection, Diauxic shift and environemntal change resonse presented with the most pronounced fold of induction or repression. In figure 2 you can see that as time in anaerobic conditions increase to 18.5 and 20.5 ERG25 shows pronounced repression (near 3 fold repression).

Figure 1 - Scale used to interpret microarray data.

Figure 2 - Yeast is exposed to anaerobic conditions and expression of ERG25 is displayed. Green=repression, red=induction
There are several genes that have similar gene expression profiles as ERG25. These genes show repression at 18.5 and 20.5 hours after start of the experiment. If you look at Figure 3 you can see that most of these genes are related to either ergosterol biosynthesis or some other biosynthesis pathway. Many of the genes in this area are unknown. Given the similarity in expression profile, one could hypothesize by "guilt by association" that these unknown genes are somehow related to either ergosterol biosynthesis or some other biosynthesis pathway.

Figure 3 - Genes with similar expression profiles under diauxic shift
When environmental conditions were changed ERG25 had several incidence of gene repression. Figure 4 highlights the three most dramatic occurances of repression in ERG25 from the Gasch AP, et al. paper. (A) shows that ERG25 is translated normally during most heat shock and cold shock conditions except one where the yeast is exposed to a temperature drop from 37 degrees to 25 degrees. This could reveal that ergostol biosynthesis is somehow important under extreme cold shock. This finding is uncertain since few of the surrounding experiments showed much repression. (B) reveals the diauxic shift experiment previously discussed. This finding leads me to believe that ERG25 and the ergostol biosynthesis pathway is someway important under low oxygen conditions. Figure 3C reveals that ERG25 in msn2/msn4 yeast is repressed when exposed to .32mM H2O2. Since this occurs in a mutant strain it is difficult to conclude anything universal about ERG25's role in toxic environmental conditions, especially since wild-type yeast had normal expression of ERG25.
(A)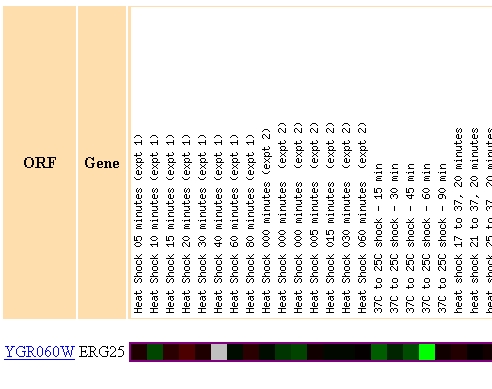
(B)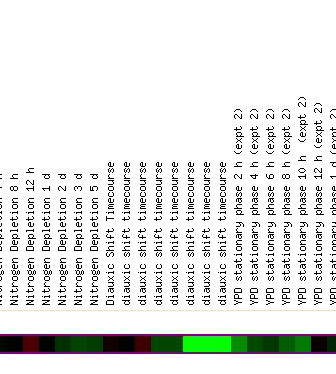
(C) 
Figure 4 - This figure shows three regions of a microarray testing the expression profile of ERG25 when yeast was exposed to varying environmental conditions. (A) Highlights a region of 3 fold repression where the yeast was exposed to 60 minutes of cold shock. (B) Highlights the diauxic shift discussed earlier in figures 2 and 3. (C) Highlights the repression of ERG25 when mutant msn2/msn4 yeast are exposed to .33mM H2O2 for 20 minutes.
When I investigated Function Juntion for more information regarding ERG25 I found some interesting findings but also one puzzling discovery. Function Junction's "Yeast Global Microarray Viewer" revealed several test conditions with high levels of induction and repression. Two findings stood out to me. When exposed to 7% ethanol for 30 minutes, ERG25 showed 2 fold induction. Thus, ERG25 and ergostol biosynthesis might have some importance when yeast cells are exposed to high levels of alcohol. It might be advantageous for the cell induce greater levels of sterol production when exposed to toxic levels of ethanol.
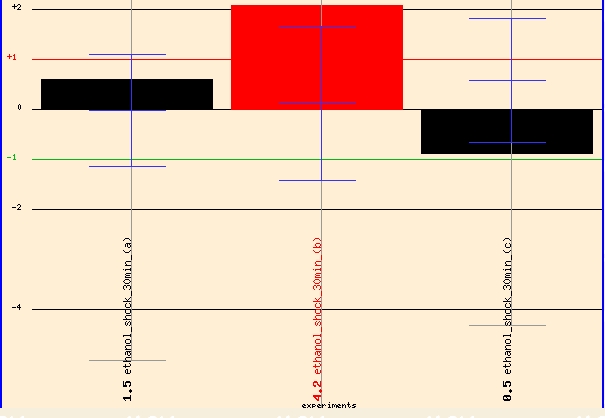
Figure 5 - ERG25 expression profile in yeast exposed to 30 minutes of 7% ethanol. (Details)
The most dramatic finding came when a group of investigators exposed yeast cells to griseofulvin. When exposed to varying concentrations of this agent for varying times, yeast cells expressed high levels of ERG25. This agent is an oral antifungal agent that has "tubulin-specific morphological effects on mitotic spindle formation" according to Savoie CJ et al's experiment (details). It appears that this agent has dramatic detrimental effects on overall yeast functioning. Thus, it presence might induce ERG25 as a reaction to a stressful and deadly agent. I couldn't hypothesize any further though on why this induction might occur. It would be interesting to investigate why when exposed to this agent the genome transcribes high levels of ERG25. How does increasing hte activity of the ergostol biosynthesis pathway
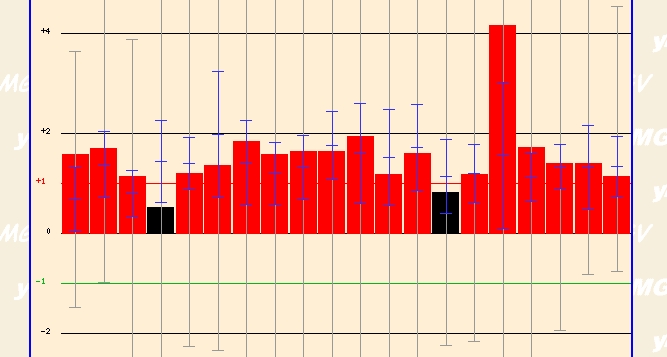
Figure 6 - ERG25 expression profile after expose to varying concentration of griseofulvin for varying times. (Details)
Another condition that revealed a dramatic change in the expression profile of ERG25 was the Yale Salinity experiment (Details) Yeast was grown in 1 m NaCl for 10, 30, and 90 minutes. The expression profile of the yeast genome was found at each time point. ERG25 had 4 fold repression after 30 minutes of exposure to 1 m NaCl. This is a dramatic level of repression that must signal that the cell is redirecting cellular functioning for other tasks other than ergostol biosynthesis--possibly as part of the environemntal stress response. At 90 minutes ERG25 has returned to its normal expression level which might signal that the cell has finished its initial environemntal stress response and has adapted to the environment; Thus the genome is restoring itself to normal expression levels.
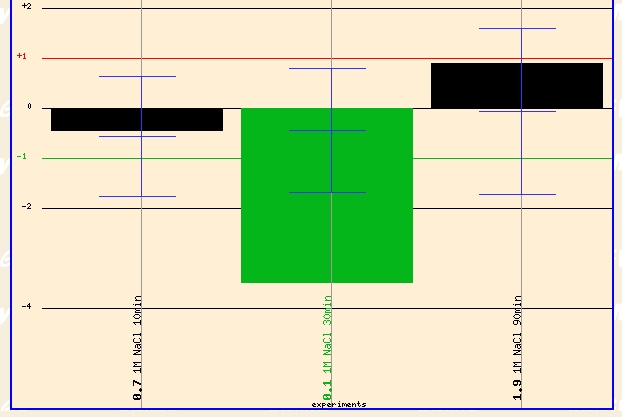
Figure 6 - Expression profile of ERG25 exposed to 1 m NaCl for 10, 30, 90 minutes. (Details)
I was puzzled by one finding at Function Junction. The site noted that there were no gene interactions (Figure 7) for ERG25. Yet from my earlier research into ERG25 I found that it had 25 related genes in the ergostol biosynthesis pathway. I stated this in my ERG25 webpage: "ERG25, or yeast methyl sterol oxidase as its gene product is called, is involved in the catalysis of the oxidation of the 4-alpha methyl group of 4,4-dimethylzymosterol. ERG25 uses ergosterol to form (22E)-ergosta-5,7,22-trien-3beta-ol, a sterol compound found in yeast and mold". Knowing that this gene is a membrane protein and interacts with other proteins in a complex process of biosynthesis, I am surprised by Function Junction's finding. I am unsure as to why they did not connect ERG25 with its related genes in the ergostol biosynthesis pathway. This requires further investigation.
Figure 7 - Function Junction states that ERG25 has no gene interactions, a finding I found strange given my earlier research
Citations
Expression Connection - http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl
Function Junction - http://db.yeastgenome.org/cgi-bin/SGD/functionJunction
Contact: Alex Trzebucki