This web page was produced as an assignment for an undergraduate course at Davidson College
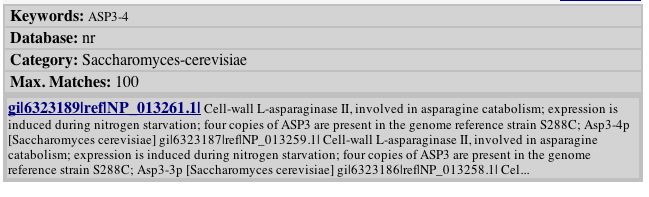
Analysis of Proteins Encoded by ASP3-4 and YLR161W
Intro
On this webpage, I will try to figure out the structure and function of the proteins encoded by ASP3-4 and YLR161w. Through searches of different proteomic databases, I will be able to categorize the role of these proteins and any connections these proteins have to other proteins in the proteome (all proteins in a cell at any one time). On my two previous yeast gene webpages, I have described the gene ontology for ASP3-4 and predicted the role of YLR161w's gene product, and this webpage will either confirm or disconfirm these results. Finally, in the absence of conclusive findings, I will describe hypothetical experiments that would help illuminate each protein's role in the proteome of yeast.
Here are the databases (and one figure) I searched for both proteins:
Benno Schwikowski Protein Connections in Yeast: PDF file
Analysis of protein encoded by ASP3-4
Nothing could be confirmed or disconfirmed as to the function of this protein through searches of the TRIPLES and ExPASy2D databases (there wasn't any data on the protein).
A search done on the PROWL database confirms the protein's role in the cell:

Fig. 1 - search results from PROWL database. Image used by permission: http://129.85.19.192/prowl-cgi/ProteinInfo.exe
A simple 2D structure found on PDB may be found here. I didn't put the picture here, because I've already put it on My Favorite Yeast Gene page.
From a search of the PROWL database, this data is given for the protein: Molecular mass: 38662.335 Da, pI: 4.7 (pI is the isoelectric point or the point at which the charge of the protien is zero. If a mixture of proteins is charged, the proteins will separate according to isoelectric points and this allows the researcher to isolate one protein in particular).
Interactions with other proteins:
In order to understand the connections specified by the DIP graphs, one must refer to the legend:
Search of the DIP database yields this graph:
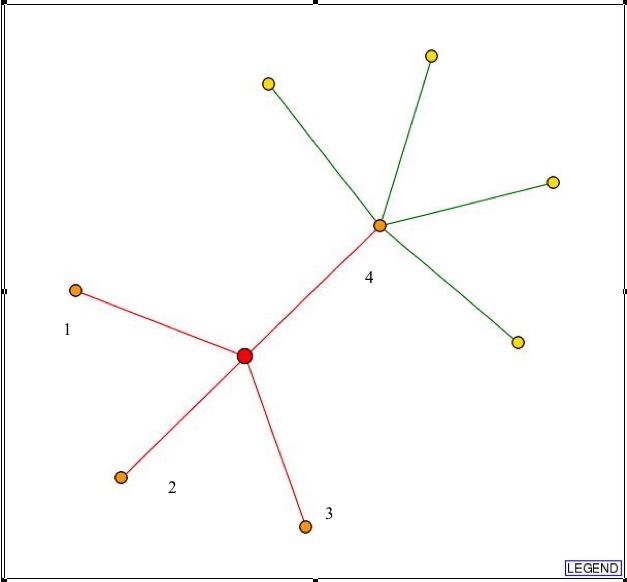
Fig. 2- Picture showing the connections L-asparaginase II has with other proteins. Picture used courtesy: http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?PK=3115
Proteins (1-4) that L-asparaginase II has connections with in Fig. 2 :
1) Tom7p:
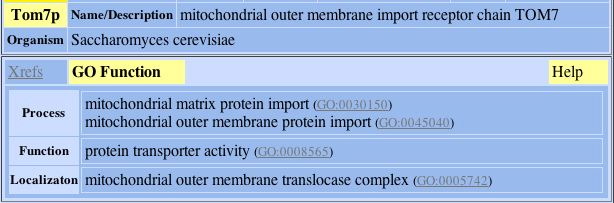
Fig. 3- Protein connection 1 with L-asparaginase II. Image used courtesy: http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?PK=4364

2)Tmt1p: not much information on how this is linked to L-asparaginase, but perhaps they are somehow linked in biological process.

Fig. 4- Protein connection 2 with L-asparaginase II. Image used courtesy: http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?PK=4807

3) Tif3p:

Fig. 5- Protein connection 3 with L-asparaginase II. Image used courtesy: http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?PK=3806
![]()
4) Atp14p:
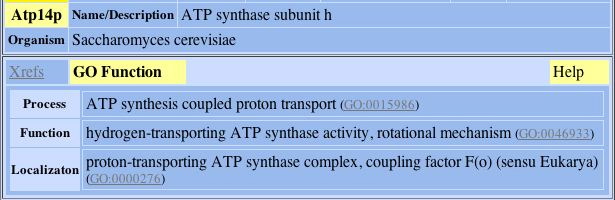
Fig. 6- Protein connection 4 with L-asparaginase II. Image used courtesy: http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?PK=3030

Now, all these proteins have a red line drawn from them to L-asparaginase II, and this means none of the connections are actually verified. So, the verification of these connections needs to be done through experiment.
ASP3-4 is not found on the Benno Schwikowski figure.
Analysis of protein encoded by YLR161w
Nothing could be confirmed or disconfirmed as to the function of this protein through searches of the TRIPLES and ExPASy2D databases (there wasn't any data on the protein).
No data on this protein from PDB, yet search of the PROWL database yields this info: Molecular mass: 13129.399 Da, pI: 9.4.
Interactions with other proteins:
From the DIP database this graph was found:
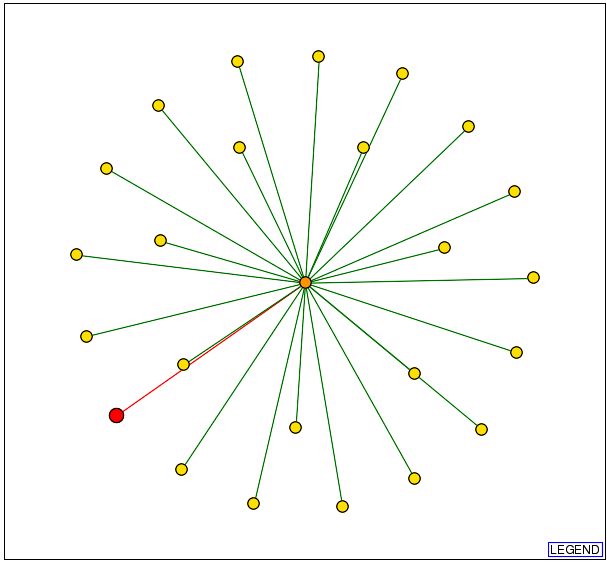
Fig. 7- This graph from DIP shows that YLR161w is connected to one protein. Image used courtesy: http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?PK=5081
The protein that the hypothetical protein of YLR161w is connected to is involved in mRNA catabolism as this image taken from DIP confirms:
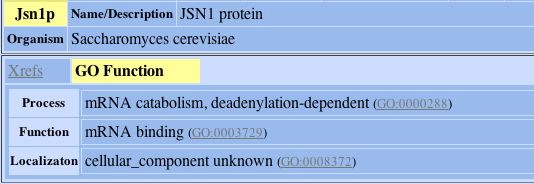
Fig. 8- Description of the one protein that YLR161w's hypothetical protein is connected to. Image used courtesy: http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?PK=5081
Protein connections found on Benno Schwikowski Figure: None
Hypothetical experiment to learn more about the YLR161w hypothetical protein
Since there is not much information on the function or structure on the YLR161w hypothetical protein I would run two simple experiments as a starting point and then, once general information is known, I would do more detailed experiments.
In order to learn more about the YLR161w hypothetical protein I would try to figure under what conditions the protein is present. In order to do this, I would utilize the method that the Snyder lab came up with, which entails using a specialized transposon (segment of DNA that can move from one location in the genome to another), called mTn, that inserts itself randomly in the yeast genome. The mTn transposon has several features that make it useful in analyzing the yeast proteome: it contains the lacZ gene which encodes an enzyme that turns a colorless substrate blue, and the gene lacks a promoter or the first codon, so the cells that turn blue have the transposon inserted into some of the genes. Also present in the sequence of the transposon is a site called 3xHA, which is an hemagglutinin (HA) epitope tag. An epitope tag is a sequence that encodes a protein that a known antibody binds to, so when a blot or microscopy is made those strains that transcribe the protein can be recognized. I would do sequence anlaysis on several strains and figure out which ones had insertions in the region of YLR161w. I would then use phenotype macroarrays to determine under what growth conditions those strains with insertions in the region of YLR161w would or would not grow in. This experiment would help to determine whether YLR161w hypothetical protein is present in normal growth conditions or whether it is a response to a certain environmental condition.
The next experiment I would run on YLR161w hypothetical protein is a yeast two-hybrid experiment to determine what proteins YLR161w hypothetical protein interacts with. In this experiment I would use YLR161w hypothetical protein as the bait and use a variety of proteins involved with mRNA as the prey. In this experiment the bait protein is bound to DNA binding domain (DBD) and the prey protein is bound to an activation domain (AD). If the two constructs interact with each other then a reporter gene, His3, is transcribed that produces an amino acid histidine.
References:
1) Dolinski, K. et al. (2003). Saccharomyces Genome Database. <http://www.yeastgenome.org/> Accessed 2004 18 Nov.
Questions or comments? email me: jobunton@davidson.edu
John Bunton's Genomics Home Page