Proteomics of LAS17 and YOR186 This web page was produced as an assignment for an undergraduate course at Davidson College. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Thus far in my studies of genomics, I have examined the DNA sequence and protein functions of one annotated and one unannotated gene in the baker's yeast (Saccharomyces cerevisae) genome. Las17 is a gene that encodes a protein that assists with actin formation. In this website, I will examine the protein characteristics and interactions of Las17. Yor186 is an unannotated gene that has no known function. In previous websites, I have used genomic techniques to predict the function of this gene. In this website, I will further examine my hypothesis using proteomics and suggest an experimental design to understand the function of Yor186. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LAS17 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Data Bank | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
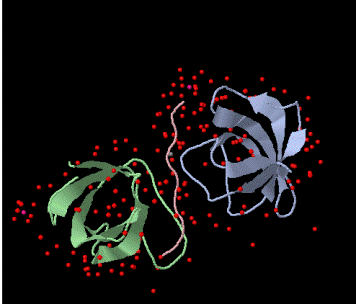
Figure 1. Schematic of the the BBC1 component of the Sh3 domain of S. cerevisae from PDB. The pink peptide chain of proline originates from LAS17. |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
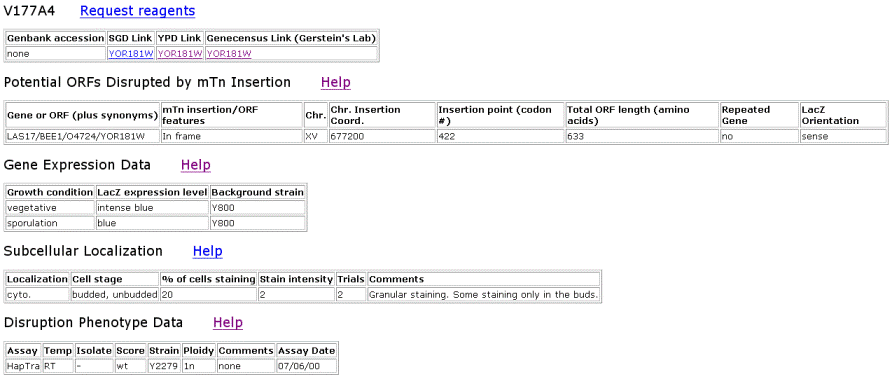
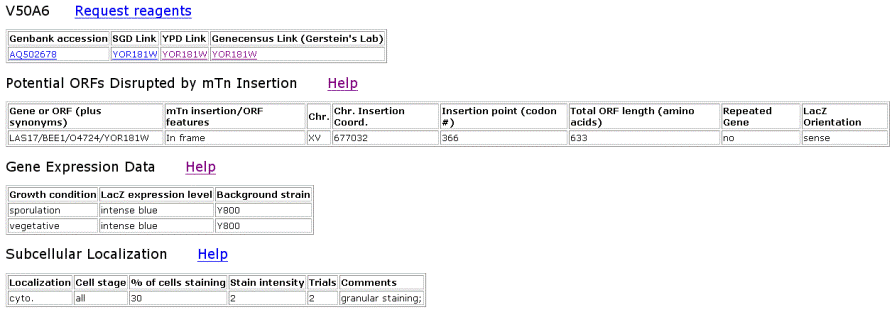
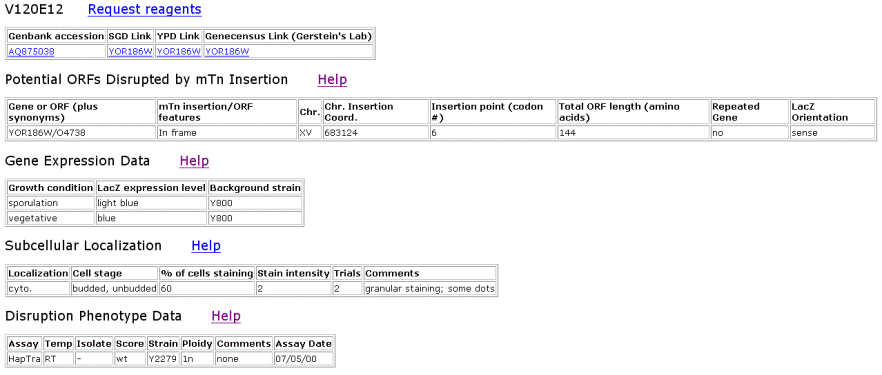
| TRIPLES Results for LAS17 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Figure 2. Data from TRIPLES that shows where the transposon mTn was inserted and how the subsequent disruption of the gene affected the cell and its phenotype. These results yield insights into subcellular locations and where the proteins are expressed. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Database of Interacting Proteins | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 |
 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
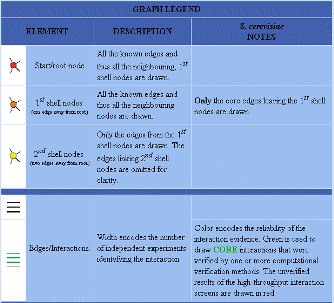
| Figure 3. Database of Interacting Proteins results for LAS17. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Interacting First Shell Nodes as Determined by the Database of Interacting Proteins | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Table 1. Proteins that interact with LAS17 during actin filament formation. Information obtained from SGD. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
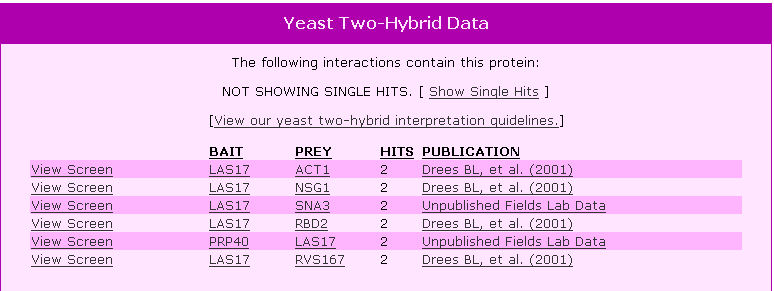
| Y2H Results for LAS17 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Figure 4. Y2H results for LAS17 using this protein as a bait and showing other proteins that LAS17 bound to as prey. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Other Databases Searched | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| YOR186w | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Facts Obtained From Protein Information Resource | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ExPasy 2-D Gel | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Figure 5. ExPasy results for YOR186w. It appears that in the black box that surrounds the area where YOR186w should be located has several small black dots indicating that low levels of protein were found there. YOR186w may or may not be represented on this gel because it is difficult to distinguish. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| TRIPLES Results for YOR186w | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Figure 6. TRIPLES results for YOR186w that shows where the transposon mTn was inserted and how the subsequent disruption of the gene affected the cell and its phenotype. These results yield insights into subcellular locations and where the proteins are expressed. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
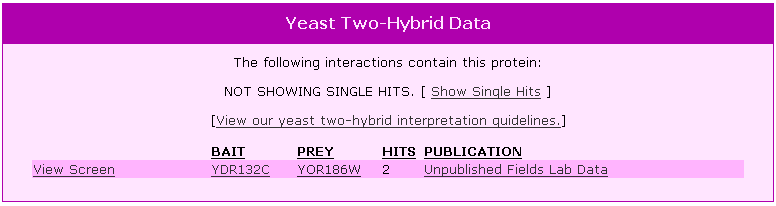
| Y2H Results for YOR186w | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Figure 7. Y2H results for YOR186w. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
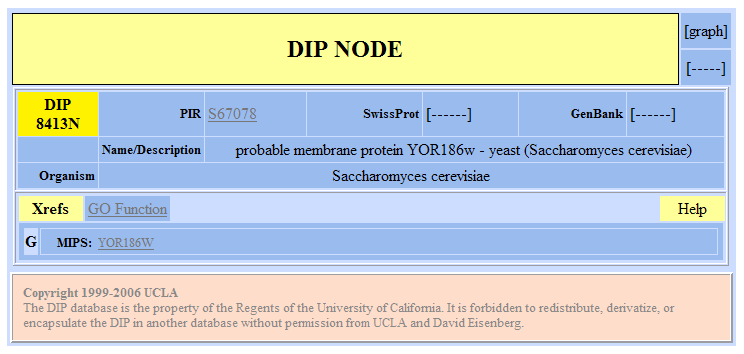
| DIP Results for YOR186w | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Figure 8. Database of Interacting Proteins results for YOR186w. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Other Databases Searched | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Conclusions of the Function of YOR186w | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Possible Experiment to Determine the Function of YOR186w in Saccharomyces cerevisae | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
My first step would be to specifically determine the cellular location of YOR186. I would try to use first immunofluorescence provided that a suitable antibody for YOR186w exists or could be developed. Next, the antibody would need to have an epitope tag that was labeled with an immunofluorescent that would glow wherever YOR186w was found within the cell. My next step would be to determine more proteins that interact with YOR186w. Techniques I could use include immunoprecipitation, crystallization, or the Y2H method. We could use immunoprecipitation to precipitate proteins that physically interact or bind with YOR186w. This requires that an antibody binds with YOR186w that when centrifuged would pull this protein and any physically bound proteins so that they may be separated out of solution. We could then identify these proteins by sequence analysis. Crystallization of YOR186w could be used to determine what molecules interact with the protein active site. We would crystallize YOR186w and then in solution add many different molecules or other proteins that fit within the active site of the protein. The final method I would recommend for identifying proteins that interact with YOR186w is the Y2H method. From the results I have found using this method, it does not appear as if YOR186w was ever used as the bait protein. Using this method, we may be able to determine which proteins interact with YOR186w. Once we have determined a location and possible protein interactions, we may be able to final deduce its function when used in conjunction with data about its DNA sequence and expression profile. Thus far, little information appears to exist about YOR186w. This is why I would encourage the use of general methods to learn more about the protein. From my predictions, I would assume that we would find that YOR186w is a nuclear membrane protein. I would also expect to find that this protein interacts with other proteins involved with DNA transcription and subsequent translation of mRNA. One method to use that would refine the prediction of YOR186w's transmembrane region, would be to use a method developed by Michel Desjardins's group at the University of Montreal. First, we would need to develop a method to separate the nuclear membrane from the cell, but if this could be accomplished, we would have data that may or may not support this prediction. We could subject the membrane to Triton X-114, a detergent that cleaves proteins at the membrane. If YOR186w was isolated from this sample, we could reasonably assume that this protein is at least bound to the membrane. We could run subsequent analyses using proteases to determine if the protein had components on either side of the membrane by cleaving proteins outside of the membrane before isolating the membrane and running the same test. If part of YOR186w is present in the results, we may assume that it is a transmembrane protein. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Databases Used | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Database of Interacting Proteins - Accessed 11/16/2006 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
ExPasy - Accessed 11/16/2006 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
KEGG - Accessed 11/16/2006 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Data Bank - Accessed 11/16/2006 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Information Resource - Accessed 11/16/2006 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
PROWL/ProFound - Accessed 11/16/2006 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Saccharomyces Genome Database - Accessed 11/16/2006 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
TRIPLES - Accessed 11/16/2006 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Y2H - Accessed 11/16/2006 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Return to Kristen Cecala's Homepage | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Questions? Please contact Krcecala@davidson.edu © Copyright 2006 Department of Biology, Davidson College, Davidson, NC 28035 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||