This web page was produced as an assignment for an undergraduate course at Davidson College.
The HAR1 gene and its role during brain development -
An Analysis of Scientific and Popular Literature
This question has been asked many times by researchers of human evolution. Like many questions in biology, the answer is probably not simple. For example, our brain is roughly three times bigger than that of chimpanzees and contains many more levels of organization than their brains, including a greater degree of asymmetry between sides of the brain (Hill and Walsh, 2005), but many of the mechanisms behind our unique brain development are unknown. One previous way to study the evolution of brain development has been to identify brain defects that lead to diseases, such as dyslexia, and determine their genetic backgrounds, thereby determining which genes control which functions in the brain (Hill and Walsh, 2005). Aside from learning about brain development, this method can also help develop better ways to combat brain disease.
Comparative Genomics Led to The Discovery of HAR1
Comparative genomics, which can study minute differences between human genomes and those of other species in both coding and non-coding regions, can provide a new way of studying human brain evolution. A recent paper published in Nature (Pollard et al., 2006) uses statistical analysis of various genomes to identify genes that changed rapidly from chimpanzees to humans, thereby providing a new method to study human evolution at the genomic level.
Around the time of the publication in Nature, multiple news articles were written, including one from USA Today, an article written for BBC News, and a Philadelphia Inquirer editorial. These three sources show different approaches taken by the news media when covering scientific discovery.
The Scientific Article -
"An RNA gene expressed during cortical development evolved rapidly in humans."
Published on August 17, 2006 in Nature (link to the abstract here; full text available by subscription)
The article begins with a discussion of the methods used to determine which genes evolved more quickly in human evolution as opposed to other times in evolutionary history. The researchers compared the chimpanzee genome to mouse and rat genomes to determine segments of sequence identity greater than 96% (Pollard et al., 2006). They aligned these sequences with segments of the human genome to determine which sequences mutated in the evolution from chimpanzees to humans. Those segments that had changed rapidly were labeled human accelerated regions (HARs). In particular, they identified a region, labeled HAR1, that significantly differed from the chimpanzee genome, displaying a total of 18 mutations over 118 base pairs as compared to a background substitution rate of 0.27 mutations in this segment of the chromosome (Pollard et al., 2006).
So what is HAR1?
HAR1 is a 118 bp region of chromosome 20 that lies in the conserved region of two different RNA genes. Both of these genes, labeled HAR1F and HAR1R, code for an mRNA but no protein.
 |
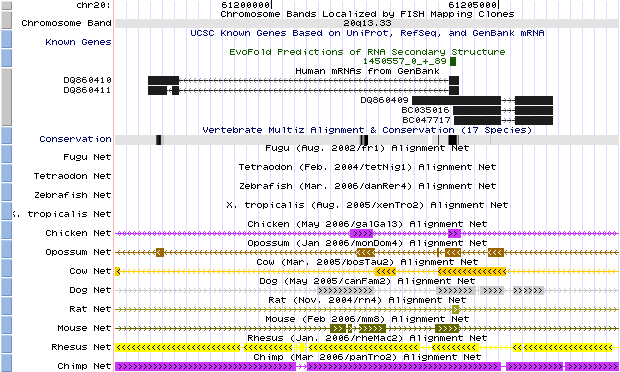
Fig. 1. Human genome browser (March 2006 assembly) description of HAR1 and its transcripts. Five human mRNAs can be transcribed from this region – the first two are splicings of a gene labeled HAR1R, while the last three are different splicings of HAR1F. According to the conservation and species comparison graphs, only a small portion of the gene, the overlap region between the two HAR1 genes, is conserved in multiple animals, pinpointing this region as the 118-bp HAR1 region (Kent et al., 2002) (http://genome.ucsc.edu/). |
The Human Genome Browser shows that there are two main mRNAs transcribed from HAR1, labeled HAR1R and HAR1F. The researchers were able to predict the structure of the conserved section of HAR1F by predicting the folding pattern of the mRNA. The paper also noted that the RNA structures of the human and chimpanzee HAR1F were structurally different (Pollard et al., 2006).
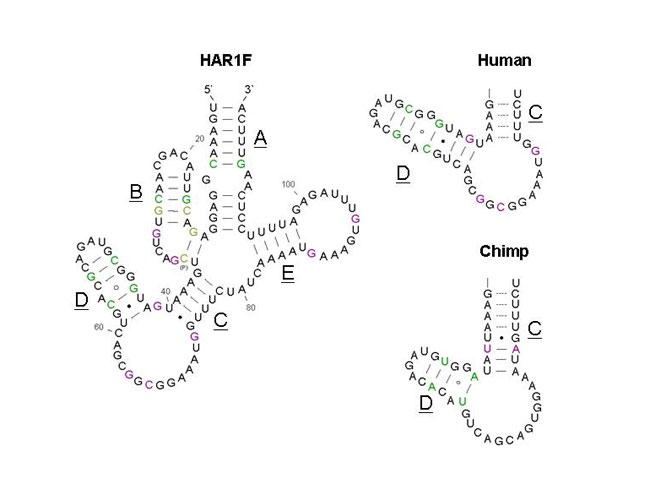 |
| Figure 2. Secondary structure of a segment of the HAR1F mRNA. On the left is the conserved section of the human HAR1F mRNA. The right two structures compare differences in the secondary structures of Human and Chimpanzee HAR1F transcripts. As seen in the picture, the lengths of chains C and D are different. (Permission Pending from http://www.livescience.com) |
Where are HAR1R and HAR1F transcribed in the body? How are they different?
To determine the location of both transcripts, researchers isolated human adult and embryonic tissue and used in situ hybridization, whereby complementary sequences of labeled RNA are inserted into tissue to locate and illuminate the target RNA. The study found that “HAR1F is strongly and specificallly expressed in the developing neocortex early in human embryonic development” (Pollard et al., 2006), specifically in cells called Cajal-Retzius neurons. These cells secrete reelin during development, which is a protein that helps coordinate proper brain structure and organization. Although HAR1F was expressed alongside reelin in the developmental stages of the brain, suggesting some interaction between the two compounds, researchers did not conclude that HAR1F influences reelin expression or vice versa (Pollard et al., 2006). In macaque monkeys, HAR1F is expressed in the same places in the developing brain, showing that the compound could have the same basic function in other vertebrates but could act differently in performing that function (Pollard et al., 2006).
The Nature article reports that in contrast to HAR1F, HAR1R is not produced in high levels until later in the developmental stages of the brain. At the end of the article, a hypothesis was given that HAR1R could help to downregulate the action of HAR1F during later parts of the brain development cycle, although research in this area is inconclusive (Pollard et al., 2006). When studying the mouse genome, the team found different levels of transcription of both HAR1F and HAR1R in the brain during development, showing that levels of transcription of these genes could have changed during evolution and could have some effect on brain development.
Were there any other interesting findings?
One of the more curious things found in the article was that “all 18 human-specific changes in [HAR1] are from A or T [weak nucleotides with 2 H-bonds] to G or C [stronger nucleotides with 3 H-bonds]. This has the effect of strengthening RNA helices in the predicted RNA structure” (Pollard et al., 2006). This finding shows that the HAR1 proteins could have become stronger through the mutations, although we can only guess how this would affect the in vivo action of the transcript.
This fact becomes even more curious when the paper continues to note that “among the 49 most accelerated [human genes], the frequency of weak to strong substitutions is 77% higher” than vice versa (Pollard et al., 2006). Again, we do not know the implications of this finding, only that it is a curious pattern in the mutations of the HAR genes.
After reading this paper, one conjectures that the HAR1 gene and its two RNA products could contribute to what makes our brain more organized than that of other animals. However, researchers were hesitant to make these conclusions in the final words of the paper. Instead they simply showed that theirs was a good method of studying HARs. By focusing on methods in their conclusions, the paper left it up to others to hypothesize on the implications of the research.
The USA Today Article -
"Scientists Find Brain Evolution Gene"
Published August 16, 2006 (Link to the article)
The USA today article simplifies the issue and changes the focus of the findings about HAR1. While the abstract of the Nature article begins and ends with a discussion of new methods to study human evolution through comparative genomics, the USA Today article begins with conclusions. The inclusion of the word ‘key’ in the opening sentence of the article embellishes the findings of the researchers. Although it draws the reader into the story, it may not be scientifically accurate.
In addition, the article simplifies, misstates, or even omits many of the findings of the Nature article. The article implies that the gene has something to do with the rapid growth of our brain instead of its complexity. The Nature article seems to conclude that HAR1 could interact with reelin, a protein that is responsible for brain organization, not brain size. Although it is possible that HAR1 deals with our brain growth, this is mentioned nowhere in the Nature article. It isn’t until the last paragraph of the USA Today article that the writer admits that the function of the HAR1 gene is unknown. How can the RNA bey a 'key' molecule if its exact function is unknown? USA Today also notes that the gene evolved “70 times faster” than the rest of our genes (Borenstein, 2006); although this approximation can be calculated by dividing the number of mutations in this gene (18) by the expected mutations (0.27), it is mentioned nowhere in the scientific paper and does not discuss the statistical significance of the findings. The article omits findings that do not lead to definite conclusions, such as those about weak-strong mutations and about two different RNA transcripts. It therefore skews the findings of the article by removing the qualifiers and intricacies of the data, simplifying it to a single conclusion about brain evolution.
A final flaw in the USA Today news story concerns its lack of discussion of methods. There is no mention of the methods that were used to determine these findings, such as in situ hybridization or computer analysis of genomes; instead, the article conjectures as to the findings of the paper without letting the reader make conclusions by his or herself. In contrast, the HAR1 paper in Nature concludes by stating that this method of studying and determining human accelerated regions is useful; even at the end it focuses on the validity of methods, not the implications of the research.
The Philadelphia Enquirer Editorial -
"Smarter than Fido"
Published August 21, 2006 (Link to the article)
While the USA Today article was printed in the news section of the paper, this article from the Philadelphia Enquirer appeared as an editorial. However, its was more true to the scientific publication than the USA Today article, mainly due to its more appropriate word choice and content. It was not without flaw, but explained the information in the Nature article without as much hyperbole.
The editorial, like the USA Today article, starts with a sensationalist catch-line, by stating that this new gene may give us “a direct glimpse into the very mechanism that makes us human” (Philadelphia Inquirer, 2006). However, the article soon states that there is not just one gene that controls brain development and that HAR1 contributes to the complexity, not just the size, of the brain. These two statements help make the editorial more similar to the Nature article, at least in language.
The editorial still makes some errors in language and focus, focusing on implications and not the reasoning behind the science. During a discussion of brain development toward the end of the article, the author rewords detailed explanations of brain development and the location of HAR1 RNA in an attempt to help non-scientists to understand the gene but also includes sentences that are unrelated to the article and seem to sensationalize the topic, such as the claim that the development of the human brain is an “explosive and decisive” issue (Philadelphia Inquirer, 2006).
The language when describing scientific aspects of the study seems to explain some concepts behind the study while ignoring other key concepts. For example, the editorial goes into detail about the expression of HAR1 but does not discuss the methods that the researchers used to determine whether this region was a human accelerated region or not. Overall, the editorial effectively communicates some of the findingds in the Nature publication using simplified language, which is one of the goals of a non-scientific publication, but tends to gloss over methods and the reasoning behind the researcher’s findings, two areas that are important parts of the original scientific article.
The BBC News Article -
"Research finds 'unique human DNA'"
Published August 17, 2006 (Link to the article)
This article, written online for BBC News, paints the Nature article in a different way from the other popular press articles. Instead of beginning with a sensational sentence, the article uses much more subdued language at the beginning, describing how scientists have found a gene that “appears to play a central role” in brain development (BBC News, 2006). The other unique aspect of the article was its focus on the methods of the researchers. Although it does not mention the statistics of the Nature article, it notes that the researchers used “sophisticated computer analysis technique[s]” (BBC News, 2006), thereby putting an emphasis on the scientists’ methods, not on their conclusions. They continue throughout the article to discuss the paper’s methods of determining where the gene was expressed, though without going into the details of in situ hybridization. In this way, the article led the reader through the reasoning of the researchers by describing what they did to get to their conclusions. By presenting a methods-focused review of the paper, avoiding the use of sensationalist language that blurs the findings of the scientific paper, and illustrating the preliminary nature of the data presented, the BBC News article provides a much more accurate source of information than the other popular press articles described above.
Discussion
These three examples of popular news media articles about a scientific paper show that news outlets often summarize, omit, and sensationalize information that comes from the scientific community. Kua reports that in a previous study, the most common errors made by the popular press when reporting on scientific papers were “omission of qualifying statements…, a lack of discussion of the methodology…, a change of emphasis…, and an overstatement of the generalizability of the data” (Kua et al., 2004). All four of these flaws can be found in the previous discussions in the popular press of the original article by Pollard, et al. However, the BBC article did present information in a more complete fashion than the others, showing that popular press articles, when written correctly, can effectively summarize a study’s methods and findings.
News articles are read by people with different interests and backgrounds, including those who may not be knowledgeable about scientific research. Accordingly, the news article must draw its readership into the article by employing exciting language. This style of reporting, however, is in contrast to the goal of scientific literature, which is to present unbiased methods and findings. These three news articles show multiple ways to reconcile the tradeoff between the two types of articles.
These articles illustrate the need to study a variety of sources in the popular press when assimilating information about scientific works. Although some articles may generalize or embellish, effective popular press articles do exist – they just may not be as easy to find.
References
BBC News. 2006 August 17. Research finds ‘unique human DNA.’ BBC News. <http://news.bbc.co.uk/2/hi/health/4797257.stm>. Accessed 9 September 2006.
Borenstein, Seth. 2006 August 16. Scientists find brain evolution gene. USA Today. <http://www.usatoday.com/tech/science/genetics/2006-08-16-brain-evolution-gene_x.htm>. Accessed 3 September 2006.
“HAR1F.” Picture. Livescience.com. <http://www.livescience.com/php/multimedia/imagedisplay/img_display.php?pic=060816_har1f_02.jpg&cap=The+human+and+chimpanzee+HAR1F+RNAs+form+stable+structures.+HAR1F+contains+a+snippet+of+DNA+which+has+evolved+much+faster+in+humans+than+chimps.+Credit%3A+Sofie+Salama+and+Jakob+Pedersen.>. Accessed 13 September 2006.
Hill, Robert Sean, and Christopher A. Walsh. 2005 September. Molecular insights into human brain evolution. Nature 437:64-6.
Kent, W.J., Sugnet, C. W., Furey, T. S., Roskin, K.M., Pringle, T. H., Zahler, A. M., and Haussler, D. 2002. The Human Genome Browser at UCSC. Genome Research. 12:996-1006.
Kua, Eunice, et al. 2004. Science in the news: a study of reporting genomics. Public Understanding of Science 13:309-22.
Philadelphia Inquirer. 2006 August 21. Smarter than Fido. Philadelphia Inquirer. <http://www.philly.com/mld/inquirer/news/editorial/15321794.htm>. Accessed 2006 September 8.
Pollard, Katherine S. et al. 2006 August. An RNA gene expressed during cortical development evolved rapidly in humans [abstract]. In Pubmed database <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=16915236&query_hl=4&itool=pubmed_docsum>. Accessed 3 September 2006.
Questions? Comments? Email brhenschen "at" davidson.edu
© 2006 Department of Biology, Davidson College, Davidson, NC 28035