Figure 1. Screen shot showing how to adjust memory on a Macintosh.
This book has many references and questions that require you to use the world wide web (WWW). The WWW was designed to use uniform standards so everyone would have equal access to all information. Unfortunately, a number of companies developed proprietary versions that do not work on all computers. In addition, new technology has been added to the original list of ways to deliver information. Therefore, this web appendix provides you with the information you need to get your computer up to speed for all the WWW sites used in Discovering Genomics, Proteomics and Bioinformatics.
Terminology
There are some basic terms that you need to know to use the links in this book.
A more complete list can be found at this location <http://www-personal.umich.edu/~zoe/Glossary.html>.
Browser - a software program that allows you to visually surf the WWW. There are two main browsers: Netscape and Internet Explorer.
URL - Uniform Resource Locator which is a technical way of saying the web address. It is the string of letters, slashes and numbers that allow your browser to see the appropriate page. With newer browsers, you no longer have to type in the "www" portion of the address, though it never hurts to add it.
Frames - When a web page is divided into sections, each section is called a frame. If you visit <http://bio.davidson.edu/courses/Molbio/websearch/SearchingNCBI.html>, you will see two frames of equal size, though the left frame has a lot of text and the right frame says "Web pages from NCBI will appear here....."
Download - to retrieve software or other computer files from a remote location to your computer. For example, you will need to download some software to see certain web page.
Java - this is a programming language that sends a small program over the WWW to run locally on your computer. Java programs are called "applets" meaning small applications or programs. Java is the major area where standards are not respected. Macintosh computers that run operating system 10 (called OS X) are reported to support all forms of Java, both the original standard and the Microsoft derivations. Mac OS 9.1 and earlier only supports the original Java standards. PCs running Microsoft windows operating systems will be able to support both the original and the Microsoft derived forms of Java.
Plug-ins - these are free software add-ons that you can download to update your browser. Plug-ins are needed for some of the newer media for delivering sound and movies.
Browsers
and Platforms
There are
two major computer platforms in
the biology world - Macintosh and PC (which stands for personal computer, also
known as IBM-compatible or Microsoft products). A new comer to the field is
Linux, and its popularity is growing among those who like to tinker and hack
with computers. Since most Linux users also run another platform and because
many plug-ins are not available for Linux, only Macintosh and PC platforms will
be addressed here.
Macintosh
Users
Macintosh
is still popular with many biologists but due to the power of Microsoft, it
has some problems interpreting pages created with Microsoft standards. Furthermore,
Mac users are a small minority of world users and so browser developers do not
always test their products on Macintosh computers. Because of these two reasons,
Mac users will probably want to download both Netscape and Internet Explorer
(often abbreviated IE).
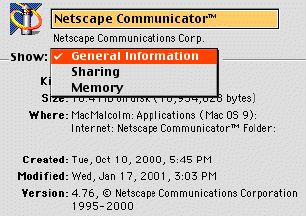
One final note about Macintoshes. As with all computers, the number of programs you can run simultaneously is determined by the amount of RAM you have bought. With Macs, you can set the RAM for each program individually so that you can have more than one open at a time even with very little RAM. However, the down side to this approach is that some big files may not open and you may get an error message. If this happens, you can fix the problem by finding the application that is RAM-limited and click on it once to highlight it but not launch it (you must quit the application if it is already running). Once the application is highlighted, hold down the Apple key and type the letter I while still holding down the apple key. This will bring up a window as shown in figure 1-1.

Figure 1. Screen shot showing how to adjust memory on a Macintosh.
Click in the box next to show: and choose memory. From this window, you can increase the allocation of RAM for any application. In this example (figure 1), the RAM is set for 40,312 K (or 40.312 megabytes). This is much higher than the default setting of 8192 and allows larger files to be viewed.
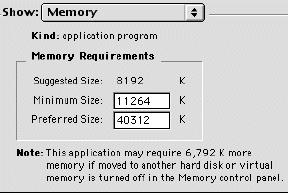
Figure 2. Screen shot showing finally settings for memory allocation.
Netscape
To download the latest
version of Netscape, go to the Netscape download home page <http://home.netscape.com/products/index.html?cp=brinavbrincs>.
As of this writing, Netscape version 6.1 is still in early form (called beta).
Since there might be some bugs (problems) with this version, this book will
assume you are using 4.x which means the most recent version of Netscape 4.
The current version is called 4.78. Download this by clicking on the link that
says "Netscape Browsers" and follow the directions. By the time the
book is published, version 6.x may be the only
option available. If so, download Netscape 6.x.
Another advantage for Netscape is the built in composer function. Netscape Composer is a free web authoring program which allows you to create your own web pages. Unless you have access to another product, you can use Composer free of charge for your web pages.
IE
The only
advantage IE has over Netscape on a Mac is Java applications. Due to Microsoft's
position in the market, it can set its own standards and expect a majority of
the world to conform. This means that only the Microsoft browser IE 5.x and
later will work with Microsoft Java applets. The newer versions of Microsoft
Java (1.1 and 1.2) may only work on Macintoshes that run with OS X. This means
in a few more years, this Java v. MS-Java conflict will fade into the distant
past.
Due to an agreement between Apple and Microsoft, IE is preloaded on newer machines. If you cannot find it, download IE by going to the Microsoft web page for browsers <http://www.microsoft.com/windows/ie/default.htm>. Click on the download button and follow directions. The current version is 5.5 and soon version 6.x will become the new standard. Download which ever is available.
PC
Users
For PC users,
Windows comes with IE built in. IE performs most functions properly. The only
exception may be chime which will be discussed below. If you need to update
your version of IE, you can go to the download page at this URL <http://www.microsoft.com/windows/ie/default.htm>.
Click on the appropriate button and follow the directions. Netscape does work
on PC's as well and you can obtain a copy from the Netscape Home Page <http://home.netscape.com/computing/download/index.html?cp=hop05hb2>.
Searching
a Web Page for a Particular Term
Here is
a simple problem with a simple solution. Have you ever searched a web page for
a particular word and had trouble finding the word after viewing the right web
site? To find the word, you can simply use the "Find" function of
your web browser and it will find the word for you. This is especially helpful
on web pages that have a lot of text.
Sample
using Find function
Go to this
URL at Cold Spring Harbor <http://www.nobel.se/chemistry/laureates/index.html>.
Up at the very top of your window, click on the "Edit" menu and choose
"Find". When a dialog box appears, type in the word "Mullis"
and hit return. You will see the word highlighted on the page. This is an easy
way to find the content you are looking for rather than having to scroll down
long pages.
Optimizing Your Browser
There are a few web sites that stand
out as places to start. We will visit a few of them here with other sites listed
at the end of this chapter.
Entrez
PubMed - http://www.ncbi.nlm.nih.gov/PubMed/
The first
place to start any project is the previously published literature. Go to the
Entrez PubMed web site to search the biomedical literature. This is run by the
National Center for Biotechnology Information <www.ncbi.nlm.nih.gov>
which is a part of the National Library of Medicine (NLM) and the National Institutes
of Health (NIH).
To access this huge database, type in any word related to biology. You will get a results page that lists all the publications that contain your word or words. The more words you use, the more specific a response you will get. If you click on the top line that has the authors names in blue, you will usually see an abstract for that publication. Occasionally there will be a large box that is a hyperlink which will take you to an online version of the original paper. The publication of science papers is experiencing a revolution of sorts and some journals allow free access to their articles immediately, others have a delay of 6 - 12 months, some never permit free access. When in doubt, click and find out.
From the Entrez page for PubMed, you can also search many other databases, In the upper left corner, there is a box that allows you to select other databases (figure 1-3). For example, you can choose to search the literature (PubMed), protein sequences, nucleotide sequences, 3D structures, whole and partial genomes, population sequence sets, OMIM which is a catalog of human health information, taxonomic definitions, and domains which are sequences that are conserved and have well characterized functions. This is the ultimate in one-stop shopping for genomic information. We will use this a lot.
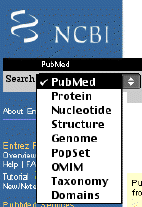
Figure 3. Screen shot of searchable databases using NCBI's Entrez web site.
Sample
Search of NCBI
Let's try
out a simple search to find a particular nucleotide sequence. Change the search
to "Nucleotide", enter the word "clock" and hit the "GO"
button. You should get a long list of hits that will cover multiple pages. Now
enter the words "fly clock"
. This should give you a very short
list. Find the one for Drosophila and
click on the accession number which is a hyperlink. You will see all the information
about this particular gene, including the protein and DNA sequences. Now change
the search to "clock Drosophila". You should get over 100 hits simply
by changing from fly to Drosophila.
Perform one last search by entering "period and Drosophila melanogaster".
You will still get many hits, even for species that are not flies because they
have descriptions that use the words you searched. Scroll down your list until
you find a sequence that says:
______________________________________________________________________________
AF251241 Protein, Related Sequences, Popset, Taxonomy
Drosophila melanogaster period (per) gene, partial cds
gi|12005687|gb|AF251241.1|AF251241[12005687]
______________________________________________________________________________
The first line has the accession number (AF251241). Below the accession number is line that describes what this hit is. The phrase "partial cds" means this is a partial coding sequence and thus in not complete. On the third line is a list of symbols that tell you a series of other accession numbers that are used in different databases for this particular sequence. On the far left side on the top line are some terms that are also hyperlinks. Click on the phrase "RelatedSequences" and you should get a short list that includes the full length sequence to the gene called period, or per for short.
Google.com
- www.google.com
If you need
to find almost any web page, the best search engine (program that finds URLs
and catalogs all relevant key words) is Google. Go to the Google web site and
you will see a small box. You may type in as many words as you want (within
reason). The more words you enter, the more specific your search will be and
Google assumes you want to find pages that include all of these terms, not one
or the other. If you know exactly what you are looking for, this is a good approach.
If you are just hunting vaguely, start with fewer terms and then add more as
you get a sense of what you are looking for.
Sample
Search
Enter the phrase
DNA microarray and very quickly you will get over 20,000 hits. You can modify
your search and add the term "undergraduate" and see that the list
has been reduced about 20 fold. You could use Google to help you find a good
summer research job.
Protein
DataBase www.rcsb.org/pdb/index.html
This protein
database (PDB) contains all computer
files that can show us the three dimensional (3D) shapes of proteins. There
are several ways to view these structures, but the easiest is to have the free
plug-in called "Chime"
which is produced by MDL (Molecular Design Limited) <http://www.mdlchime.com/chime/>.
You will have to register to get your free copy of the plug-in. Once you have
logged in, you can follow the links to the download page. It works on both Mac
and PC so choose the appropriate one. Once you have installed it, you will need
to restart your browser so the new plug-in can become activated.
Now that you have downloaded the chime plug-in, you are ready to see 3D structures that have file names ending in ".pdb". If you know the PDB file name, you can enter it in the box. If you do not know the PDB ID number, you can use words to search the database (figure 1-4). Using the PDB ID, enter 1AI3, select the "query by PDB id only" box, and click on the "Find a Structure" button. You will see a page that describes isocitrate dehydrogenase (IDH).
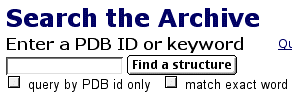
Figure 4. Screen shot from PDB web site.
A new browser window will appear. In this window, you will see the amino acid sequence for IDH in the top frame and the structure in the bottom right frame. Don't rotate the protein yet, leave it in its original position. Click on the button at the left, half way down, that says "Secondary Structure". You will see that the amino acids that make up alpha helices are highlighted in red, beta pleated sheets in blue, and bends in yellow. This has occurred in the amino acid sequence as well as the structure.
If you place your mouse over any amino acid in the structure diagram, you will see its has been identified in the black window on the top left side, just under the full sequence. This also happens when you mouse over amino acids in the sequence.
Change from "Secondary Structure" to "Exposure". You will see that amino acids on the surface of the protein are highlighted differently from the rest of the protein. Note the color of the first two amino acids (ME) in the sequence at the top. Using the mouse, find the first amino acid of the protein structure; it is located at the bottom center of the structure frame. Which amino acid is first in the structure? What happened to the first two amino acids?
Finally, click on the reset button at the bottom on the left side. Change the color to yellow. Now use your mouse to find the amino acid sequence YICLRPVRYYQ which begins at amino acid 125 and ends at number 135. Click and drag to highlight these 11 amino acids and notice that this portion of the structure has also been highlighted yellow.
Close the Quick PDB window and you should still have the original page for viewing IDH. Click on "First Glance" and an animated version of IDH should appear. You can choose to turn on and off the different options by clicking on the appropriate boxes.
Now go back one page and click on the "Protein Explorer" button. Next, make sure your window is properly sized and then click on the button to view 1AI3 from the PDB server. Although it takes a while to load, do not do anything until you see a spinning model of IDH. In the upper right frame, you will see a link that says "Explore 1AI3". Click on this and wait until you see a green box that says ready appear below the structure of IDH. A new set of buttons will appear in the top right frame. Click once on the one that says "water" and most of the red balls will turn to spheres of dots. Click again and they disappear. Click on the other buttons to see what happens.
Finally, there are a number of people who have collected some wonderful tutorials on particular molecules. If you want to visit some, try these out to see what can be done with chime scripting.
Other PDB Sites
Protein Explorer- http://www.proteinexplorer.org/
This site is maintained by Eric Martz at the University of Massachusetts who has pushed Chime scripting further than anyone else. Martz has tutorials on using Protein Explorer, How to create chime scripts, and has many tutorials for your edification.Online Molecular Museum - www.clunet.edu/BioDev/omm/gallery.htm
This site is maintained by David Marcey at California Lutheran University. Marcey and his students have created some outstanding tutorials. Click on the link at the bottom of the left side that says "the exhibits".Nucleic Acid Database atlas - http://ndbserver.Rutgers.edu/NDB/ndb.html
This database contains DNA, RNA, protein-nucleic acid structures. This may be useful if you want to look at non-protein structures.
QuickTime
(QT) - http://www.apple.com/quicktime/download/
QuickTime
is a free plug-in that allows you to see movie files. The 15 second biographies
that are a part of the online resources for this textbook utilize the QT plug-in.
The latest version of QT is 5.x and can be downloaded for Macintosh and PC computers
from the Apple web site listed above. Provide the information, choose your platform,
and download.
Sample
Movie
To make
sure your QT is working, you can check out a 15 second biography <http://bio.davidson.edu/courses/genomics/15secbios/15secbios.html>.
Choose your favorite topic and then select a biography to see and hear. You
can stop the movie by using the control buttons.
If the movies do not play properly, then you will need to check your preferences. To do this, choose preferences under the edit menu. Select "Applications" from the list of preferences. You will get a new dialog box; scroll down until you see "MPEG media file" or similar description. Select this line by clicking on it once and then click on the edit button. Make sure the button next to "Plug-in" has been selected and then make sure the most recent version of QuickTime (5.0.2 or greater) appears in the pop-up menu. If it does not, then you will need to select it by searching through your hard drive and locating QuickTime.
Flash
Animations - http://www.macromedia.com/downloads/
Flash is
the software that creates animations for the WWW, TV, movies. It is a very powerful
program that is sold by Macromedia. The plug-in is free and you can download
it from the site above. You will want to choose the option that says "Macromedia
Shockwave Player". Click on this link and follow the directions. It works
for PC and Mac, Netscape and IE.
Sample
Animation
There are
many good educational animations that use Flash. Some are included with this
book. Try out this one that describes how immunoprecipitations are performed.
This is used for one case study in Chapter 2 <http://bio.davidson.edu/courses/genomics/IMPfolder/IMP.html>.
This animation includes sounds so if you are viewing this where it is
OK to turn up the sound, do so now or use headphones. If you are in a library,
you might want to click on the link at the bottom left that will take you to
a silent version.
Adobe
Acrobat Reader - http://www.adobe.com/products/acrobat/readstep.html
Adobe is a software company
that makes a program called Acrobat. Acrobat will convert any text file into
a ".pdf" format
that stands for Portable Document File. Most browsers come with Acrobat Reader
free plug-in, but if you cannot read see a PDF file, then you can download it
from the page above. Be sure to select the free Reader program and not the full
conversion program that costs about $250.
Sample
PDF
Go to PubMed
< www.ncbi.nlm.nih.gov/PubMed/
> and enter these three authors " Evans Skrzynia Burke". You should
get one hit entitled "The complexities of predictive genetic testing".
Click on the hyperlink of the authors' names and the resulting page has the
abstract. Above the title is a box that hyperlinks to the original paper at
the journal's web site. Click on the box and you will see an html version of
the paper. In the upper right hand corner is a link that says "PDF of this
article". Click on this and then click on the "Download" hyperlink
that appears in a small box. This box gives you a short citation for the paper
and tells you the size of the file you are about to download (217K = 217 kilobytes).
Click on the download link and your browser will launch the Acrobat Reader plug-in
so you can see the paper as it appeared in the original journal. It is a very
good paper if you want to read up on this topic.
There are many other good research papers that are freely available at PubMed Central <http://www.pubmedcentral.nih.gov/> which is funded by your tax dollars and another set is available at HighWire Press <http://highwire.stanford.edu/> which is a commercial provider. You can search these two sites for many excellent journals that serve papers in Acrobat format.
Java
- platform specific links
Macintosh - http://www.apple.com/java/
PC - http://www.microsoft.com/java/
As noted above in the definitions, Java is not as universal as it could have been. You will need to got to the appropriate platform link and download the latest Virtual Runtime Machine. Make sure you match your platform, operating system, and virtual runtime machine. Macintoshes tend to work better with IE than Netscape versions 4.7x. As of this writing, Netscape 6.x was still in beta version and was not tested. If you are running a Macintosh on
OS X, you might not have any problems with Java developed by the original standards, or Microsoft standards.
Sample
Java
If you go to the SNP Consortium's database, they make nice use of Java.
Go to this URL <http://snp.cshl.org/db/snp/snp?name=TSC0019265>
and scroll down to the link that says "View Traces":
Click on this link and look at the DNA sequences for these particular single nucleotide polymorphisms. You can click on any of the options and use the scroll bar to view the entire sequence.
Web authoring (free via Netscape Composer)
One reason to keep using Netscape instead
of IE is that Netscape comes with a program that allows you to create your
own web pages - Netscape Composer. If you need to create web pages for your
course work, you can use these links.
A WWW Template for you to use
bio.davidson.edu/courses/genomics/webauthor/template.html
bio.davidson.edu/courses/Molbio/dreamweaver/dreamweaver.html
How
to use Netscape Navigator to Edit your Web Page
bio.davidson.edu/courses/genomics/webauthor/Goldediting/CommuniEdit.html
How
to make Greek Letters
/bio.davidson.edu/courses/genomics/webauthor/webfolder/Greekletters.html
How
to add sounds to your web pages
pratt.edu/~cg520/web_sounds/audio.html
How
to Create Relative Links for your Web Pages
bio.davidson.edu/courses/genomics/webauthor/webfolder/Relativelinks.html
How
to Insert a Chime image in your Web Page
bio.davidson.edu/courses/genomics/webauthor/webfolder/EmbedChime.html
Related Links
How
to evaluate a WWW site
http://bio.davidson.edu/courses/genomics/webauthor/evaluate.html
Web
Standards for this Course
http://bio.davidson.edu/courses/genomics/GPBwebstandards.html
Additional Online References
Automated
Literature Searches via PubCrawler
http://www.gen.tcd.ie/pubcrawler/
You can use this feature of PubMed to be notified of any publications
that fit a description of your design. This is a great way to stay on top
of all the developments in your field of interest.
Bioinformatics Dictionaries
Andrew
C.R. Martin – Lecturer at Reading College, UK
http://sapc34.rdg.ac.uk/~andrew/dictionary/
Human
Genome Project Glossary
http://www.ornl.gov/TechResources/Human_Genome/glossary/
SGD
Glossary
http://genome-www.stanford.edu/Saccharomyces/help/glossary.html
Medical dictionaries
Online
Medical Dictionary
http://cancerweb.ncl.ac.uk/omd/help.html
MedTerms.com
http://www.MedTerms.com/script/main/AlphaIdx.asp?p=A_DICT
Alphabetical listing of many medical terms. You can choose a letter and
browse, or enter a term and search.
Merriam
Webster Medical Dictionary
http://www.intelihealth.com/IH/ihtIH?t=9276&p=~br,IHW%7C~st,408%7C~r,WSIHW000%7C~b,*%7C
Multilingual
Glossary of technical and popular medical terms in nine European Languages
http://allserv.rug.ac.be/~rvdstich/eugloss/welcome.html
CBS
HealthWatch
http://healthwatch.medscape.com/medscape/p/gcommunity/ghome.asp
(searchable drugs, diseases, terms)
DrKoop.com
http://www.drkoop.com/
The
Lightning Hypertext of Disease
http://www.pathinfo.com/index.htm
Cell Biology Terms
Cell
Biology Dictionary
http://www.mblab.gla.ac.uk/~julian/Dict.html
Hyper
Textbook
http://esg-www.mit.edu:8001/esgbio/chapters.html
Human
Genome Glossary of Genetic Terms
http://www.nhgri.nih.gov/DIR/VIP/Glossary/pub_glossary.cgi
Student Guide
to the Human Genome Project
http://www.ornl.gov/hgmis/education/students.html
NOVA
program: Cracking the Code of Life
http://www.pbs.org/wgbh/nova/genome/
Good animations and you can watch the entire program by streaming video,
free of charge.
Pharmacology Web Pages
About.com
(legal drug information)
http://pharmacology.about.com/health/pharmacology/mbody.htm
Food
and Drug Administration Drug Information (includes drugs being evaluated)
http://www.fda.gov/cder/drug/default.htm
RxList.com
http://www.rxlist.com/
The
Access Project list of AIDS medications
http://www.aidsinfonyc.org/network/access/drugs/index.html
© Copyright 2002 Department of Biology, Davidson College, Davidson,
NC 28036
Send comments, questions, and suggestions to: macampbell@davidson.edu