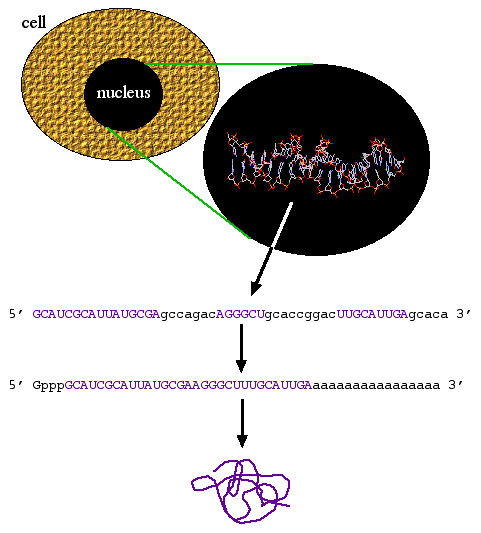
Central dogma states that biological information goes from DNA to RNA to protein (figure 1). However, there are times when information goes from RNA to DNA. Viruses such as HIV have RNA genomes that can be converted into DNA by an enzyme called reverse transcriptase. Molecular biologists realized that they could use reverse transcriptase to convert mRNA into complementary DNA and thus was born the term cDNA.

Figure 1. Central dogma: DNA to RNA to mRNA to protein. Coding sequence (purple) exons are spliced together and the 5' cap and 3' polyA tail is added to produce a mature mRNA molecule from the primary transcript. The mRNA is translated into protein.
cDNA is a more convenient way to work with the coding sequence than mRNA because RNA is very easily degraded by omnipresent RNases. This the main reason cDNA is sequenced rather than mRNA. Likewise, investigators conducting DNA microarrays often convert the mRNA into cDNA in order to produce their probes. Let's see what is required to produce cDNA.
By definition, cDNA is double-stranded DNA that was derived from mRNA which can be obtained from prokaryotes or eukaryotes. Once the mRNA is isolated, you need a few more reagents: dNTPs (dGTP, dCTP, dATP and dTTP), primers, and reverse transcriptase which is a DNA polymerase (figure 2). Mix the mRNA with the other reagents and allow the polymerase to make a complementary strand of DNA (first strand synthesis). Next, the mRNA must be removed and the second strand of DNA synthesized. There are many technical details in these steps, but we do not need to focus on them at this time.
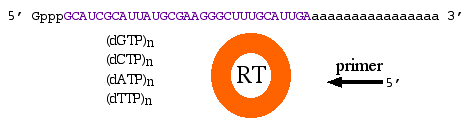
Figure 2. Four basic reagents needed to produce cDNA: mRNA as template, dNTPs, reverse transcriptase and primers.
The only issue worth mentioning now is that three different types of primers can be used (figure 3). 1) If the mRNA has a poly-A 3' tail, then an oligo-dT primer can be used to prime all mRNAs simultaneously. 2) If you only wanted to produce cDNA from a subset of all mRNA, then a sequence-specific primer could be used that wil only bind to one mRNA sequence. 3) If you wanted to produce pieces of cDNA that were scattered all over the mRNA, then you could use a random primer cocktail that would produce cDNA from all mRNAs but the cDNAs would not be full length. The major benefits to random priming are the production of shorter cDNA fragments and increasing the probability that 5' ends of the mRNA would be converted to cDNA. Because reverse transcriptase does not usually reach the 5' end of long mRNAs, random primers can be beneficial.
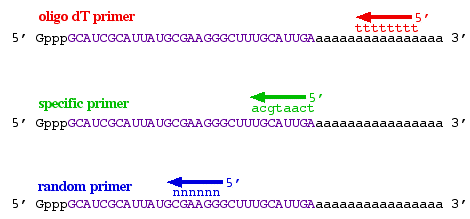
Figure 3. Three ways to prime the production of cDNA: oligo-dT primer (red), sequence-specific primer (green), random primer (blue).
© Copyright 2002 Department of Biology, Davidson College, Davidson, NC 28036
Send comments, questions, and suggestions to: macampbell@davidson.edu