General Overview:
The ubiquitous transcription factor NFAT (nuclear factor of activated T-cells)
associates with other proteins to bind DNA and induce genes responsible for
cell-cell interactions (Crabtree et al., 2002). NFAT is expressed in a variety
of lymphocytes: T cells, B cells, natural killer (NK) cells, monocytes and non-immune
related cells: muscle, cardiac, and neuronal (Scott et al., 2001, Janeway et
al., 2001). In the literature there are 4 reported NFAT isoforms, which are
designated as NFAT1, NFAT2, NFAT3, NFAT4 (Masuda et al., 1998). The presence
and redundancy of these homologs suggests the critical nature of NFAT expression
in immune and non-immune related cells. NFAT transcriptional activity is modulated
by cytoplasmic Ca2+ concentration through various Ca2+ associated signaling
pathways (Masuda et al., 1998, Crabtree et al., 2002). Increases in cytoplasmic
Ca2+ concentration induce NFAT dephosphorylation and NFAT translocation to the
nucleus where it binds to cis regulatory elements of target genes as a monomer
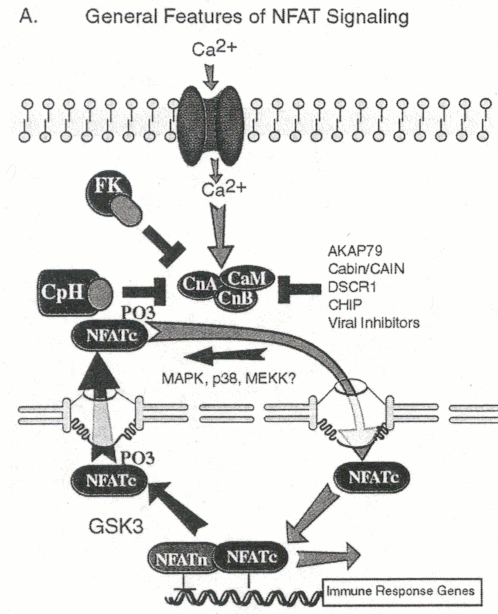
(Masuda E.S. et al., 1998). Figure 1 outlines a general signaling mechanism
for NFAT activation.

Figure 1: General signaling pathway for NFAT. ATP hydrolysis is not included in the cartoon (Crabtree et al., 2002) Permission Pending for image use.
NFAT Structure:
NFAT secondary structure consists of b sheets and random coils forming three
functional domains: a Rel-similarity domain (RSD) for DNA binding and association
with AP-1 transcription factors, a NFAT homolog region (NHR) domain, containing
intracellular localization signaling sequences, and transcriptional activation
domains (TAD), which recruit coactivators of NFAT transcription CBP, p300 and
JAB1 after NFAT:AP-1 protein complex formation (Crabtree et al., 2002). Phosphorylating
and dephosphorylating the NHR domain determines intracellular localization.
NHR phosphorylation results in nuclear export and cytoplasmic localization and
NHR dephosphorylation results in nuclear import and nuclear localization. NFAT
also contains two sequence motifs: a nuclear location signal sequence (NLS)
and a nuclear export signal sequence (NES) that permit the import and export
of NFAT from the nucleus, respectively (Janeway C. A. et al., 2001). NFAT contains
two NLS motifs, one in the NHR domain and one in the RSD domain, the latter,
however, is less efficient at translocating NFAT to the nucleus. Also mutations
in the basic NLS domain reduce NFAT translocation. (Masuda et al., 1998)
The tertiary structure of NFAT has two domains that resemble immunoglobulin
(Ig) folds. One domain, in the RSD, specifically binds to DNA and is a beta
barrel comprised of three beta sheets. Within the beta barrel there are two
prominent loop structures, the first loop (designated as the DNA binding loop)
is more constrained and binds to the DNA major grove at the 5’ recognition
sequence GGAAAA. The second more mobile loop (designated as the Rel-insert region)
binds to the minor grove at the 3’ recognition sequence GGAAAA. (Masuda
et al., 1998) The anatomical structure, topology and protein interactions of
NFAT are depicted in figures 2 and 3.
Figure 2: A 2.7 Angstrom resolved crystal structure of NFAT, Ap-1 heterodimer (Fos:Jun), and a distal portion of the IL-2 gene promoter (Chen et al., 1998). Image from PDB.
Figure 3: Cartoon of NFAT crystallized with AP-1 proteins and DNA. The DNA promoter bound to the NFAT, AP-1 protein complex is ARRE2, part of the IL-2 promoter (Chen et al., 1998). The protein-protein interactions for this complex are largely polar and hydrogen bonding and salt bridges cooperatively link the protein-protein-DNA complex. (Szilak et al., 1999) These extensive interactions permit the bending of Fos and DNA to form an extended grove which interacts with 15 DNA base pairs (Chen et al., 1998). Note the Rel-homology region (RHR) N domain of NFAT extensively interacts with the Fos-Jun heterodimer. As Jun moves away from NFAT, Fos’s luecine zipper moiety interacts with the NFAT RHR-N domain (Szilak et al., 1999). Premission pending for image use.
A mechanism for NFAT regulation: Ca2+ signalling via
the TCR leads to NFAT activation:
Two enzymes, calcineurin, a cytoplasmic serine/threonine phosphotase and nuclear
glycogen synthase kinase 3 (GSK3) are responsible for the regulation of NFAT
transcriptional activity. Calineurin upregulates NFAT by dephosphorylating serines
in the SP-repeats and in the serine rich N terminus region of NFAT (Crabtree
et al., 2002). This induces a conformational change exposing the NLS (Scott
et al., 2001), allowing NFAT to translocate to the nucleus. Calcineurin activity
is regulated by cytoplasmic Ca2+ concentration. In T cells, TCR stimulation
spurs a biphasic increase in intracellular Ca2+ concentration. The initial wave
of Ca2+ concentration from Ca2+ sequestered ions in the ER, the second results
from an influx of extra-cellular calcium ions from Ca2+ release activated Ca2+
channels (CRAC) (Crabtree G. R. et al., 2002). CRAC activation is required for
upregulation of NFAT controlled genes since transient increases in Ca2+ concentration
from the ER is not sufficient to promote NFAT activation (Crabtree et al., 2002).
This indirectly suggests that translocated NFAT must overwhelm GSK3 in the nucleus
in order for NFAT to bind to target genes.
GSK3, on the other hand, downregulates NFAT by phosphorylating NFAT and inducing
a conformation change exposing the NES (Masuda et al., 1998).
NFAT regulates….
NFAT activation regulates a variety of immune processes: apoptosis, anergy,
T-cell development, and ageing of immune system. In particular, NFAT isoforms
are responsible for regulating interluekin (IL) 2, IL-3, IL-4, IL-5, granulocyte
macrophage colony-stimulation factor (GM-CSF), interferon (IFN)-y, tumor necrosis
factor (TNF) alpha and the cell surface receptors CD40L, CTLA-4 and FasL expression
(Masuda et al., 1998). Macian et al. further characterized NFAT regulatory roles,
hypothesizing that transcriptional activity mediates two distinct gene expression
patterns which are independent and dependent upon NFAT:AP1 interactions. NFAT1
mutagenesis studies demonstrated that NFAT:Ap-1 complexes are required for inducing
IL-2, GM-CSF, IL-3, IL-4, MIP1 alpha, and FasL. Conversely, the expression of
TNF alpha and IL13 promoter activity does not require NFAT:Ap-1 interactions
(Macian et al. 2002). The NFAT:Ap-1 complex is also required to induce NFAT
activated cell death. (Macian et al. 2000) Additional regulatory roles have
been observed for NFAT. For example, Gomez et al. reported that NFAT translocation
regulates expression of the anti-apoptotic protein Bcl-2. Inhibition of calcineurin
downregulates Bcl-2 expression in IL-2 cells. Conversely, constitutive calcineurin
expression upregulates Bcl-2 expression, indirectly implicating the function
of NFAT in Bcl-2 expression (Gomez J. et al., 1998). Sheng Xiao et al. observed
that FasL transcription could be induced by two different promoters which are
mediated by NFAT binding. Only one promoter, however, required Sp1 and NFAT
binding. Thus, the aforementioned studies suggest that NFAT has a variety of
regulatory roles in cells that are dependent upon promoter and NFAT:protein
complexes.
Receptors inducing NFAT activity:
The most prominent and studied NFAT activation pathway is through the TCR. TCR
stimulation by MHC molecules, in conjunction with CD28:B7 costimulatory signal
induces a biphasic intracellular Ca2+ influx that induces NFAT dephosphorylation
and translocation to the nucleus (Janeway et al., 2001). In B cells, BCR stimulation
or CD40 and IL-4R stimulation induces NFAT activation. Stimulation of FceR1
in mast and basophil, and FcgRIII in NK cells also induces NFAT dephosphorylation
and translocation, respectively. (Masuda et al., 1998)
Inhibitors of NFAT:
NFAT inhibitors can be divided into two class, protein inhibitors and small
molecule inhibitors. Most of these inhibitors bind calcineurin and suppress
dephosphorylating activity. To date, there are four protein inhibitors which
prevent NFAT nuclear translocation: AKAP79, a scaffold protein that prevents
calcineurin substrate interactions, CABIN protein, which blocks calcineurin
activity, a calcineurin B homolog, CHP, and MCIP1,2,3 proteins which have the
ability to prevent NFAT2 phosphorylation and nuclear import (Crabtree et al.,
2002).
The two most prominent NFAT small molecule inhibitors are cyclosporin A and
FK506. Mechanistically, cyclosporin A and FK506 indirectly repress NFAT by inhibiting
calcineurin activity. These drugs target NFAT specific pathways and act as immunosuppressants
by inhibiting alloreactive T-cells. Clinically, these drugs are administered
to patients to prevent graft reject (Janeway C. A. et al., 2001) and have reduced
joint erosion and disease progression, yet there are also side effects: nephrotoxicity,
neurotoxicity, diabetogenicity, and gastrointestinal toxicity (Trevillyan et
al., 2001). Several new drug candidates, 3,5-bistriflouromethyl pyrazole (BTP)
derivatives inhibit Th1 and Th2 cytokine gene expression thereby indirectly
inhibiting the nuclear localization of NFAT. These BTP derivates, however, do
not dephosphorylate calcineurin and a mechanistic model for inhibition of NFAT
pathways has not been elucidated (Trevillyan J. M. et al. 2001).
NFAT knockout mice:
Several studies have been conducted using knockout NFAT mice to determine NFAT’s
functional role in the immune and non-immune related cells. Masuda et al. reports
that NFAT1 (-/-) mice have deregulated transcription of certain genes accompanied
by the hyperproliferation of splenic B and T cells. This hyperproliferation
is due to a lack of FasL expression which would normally induce cell death in
surviving cells (Crabtree et al., 2002) In T-cells stimulated with anti-CD3
there was noticeably reduced IL-4 transcription. Hence, Masuda et al, posits
that NFAT1 is a positive and negative regulator of cytokine expression. NFAT4
is also highly expressed in CD4+CD8+ thymocytes and NFAT4(-/-) mice have reduced
CD4+CD8+ cell counts (Amaskaki et al., 2002). Crabtree et al. suggests that
the loss of double positive thymocytes results from Bcl-2 suppression during
thymic development. Mice with mutant NFAT1/4 undergo spontaneous differentiation
in to T helper 2 cells with excess IgE production resulting in strong allergic
responses. These mice also have spontaneous T-cell hyperproliferation, which
does not depend on CD28 stimulation for activation (Crabtree et al. 2002).
Subsequent studies with knockout mice have revealed the additional importance
of NFAT activity in cell differentiation and development. NFAT1 (-/-) mice also
have small-multinucleated muscle cells, while older mice have excess cartilage
resulting in decreased joint mobility. Horsley et al. observed that NFAT2 (-/-)
mice have “defective heart valve development and abnormalities in the
cardiac septum”. In NFAT3/4 double knockout mice, embryo fatality result
from disrupted blood vessel organization and instability in the vessel walls.
NFAT4 knockout mice have defective embryonic myofibers (Horsley V. et al, 2002)
NFAT and T-cell’s:
NFAT expression and trancscriptional activity are critical for the proliferation
and differentiation of armed effector T-cells (Janaway et al. 2001). NFAT has
also been implicated not only in inducing genes for an immune response but also
a set of anergy associated genes. Weak TCR and BCR stimulation can induce NFAT
activation by low sustained intracellular Ca2+ concentrations. Macian et al
observed that null T cells for NFAT1 do not undergo anergy and show lower expression
levels of anergy-genes. For T cells with “a constitutively active NFAT1”
where AP-1 is not induced, or NFAT:AP-1 interactions do not occur had increased
expression of anergy related genes indicative of an anergy associated phenotype
with lower TCR responsiveness (Macian et al. 2002).
NFAT and Disease:
The ubiquitous nature of NFAT expression and its regulatory role in cell differentiation
and development makes it a critical enzyme in adaptive immune responses. In
patients with defective T-cell proliferation and IL-2 synthesis, researchers
noticed detectable Ca2+ release from the ER where as Ca2+ release from the plasma
membrane was not detected. This data supports the hypothesis that a sustained
intracellular calcium concentration is required for NFAT translocation via the
calcineurin GSK3 signaling pathway (Masuda et al, 1998).
Various viral infections also positively and negatively interfere with NFAT
signaling pathways. Scott et al. observed that GFP-NFAT nuclear translocation
was blocked in the early stages of herpes simplex virus (HSV) infection. HIV
hepatitis C and African swine fever are also believed to interfere and repress
NFAT transcriptional activity. In particular, the protein A238L of African swine
fever virus binds to calcineurin, thereby blocking NFAT translocation (Crabtree
et al., 2002). Kaposi sarcoma associated herpes virus and rhesus monkey rhadinoviruss,
which, infect B cells, however, stimulate NFAT transcriptional pathways and
may be responsible for lymphoproliferative disorders (Scott et al. 2001).
References:
Janeway C. A., Travers P., Walport M., Shlomchik M. J. 2001. Immunobiology.
New York, NY: Garland Publishing. p. 204-205
Masuda E. S., Imamura R., Amasaki Y, Arai K, Arai N. 1998. Signalling into the T-Cell Nucleus: NFAT regulation. Cell Signal. 10 (9): 599-611.
Xiao S, Matsui K., Fine A., Zhu B., Marshak-Rothstein A., Widom R. L., Ju S-T. 1999. FasL promoter activation by IL-2 through SP1 and NFAT but not Egr-2 and Egr-3. Eur. J. Immunol. 29:3456-3465.
Szilak L., Moll J. R., Vinson C. 1999. Structure of the DNA-Binding Domains from NFAT, FOS and JUN Bound Specifically to DNA. Chemtracts. 12: 768-773
Trevillyan J. M., Chiou X. G., Chen Y. W., Ballarom S. J., Sheets M. P., Smith M. L., Wiedeman P. E., Warrior U., Wilkins J., Gubbins E. J., Gagne G. D., Fagerland J., Carter G. W., Jay R. L., Mollison K. W., Djuric S. W. 2001 December 21. Potent Inhibition of NFAT activation and T Cell Cytokine Production by Novel Low Molecular Weight Pyrazole Compounds. Journal of Biological Chemistry 276(51): 48118-48126.
Scott E. S., Malcomber S., O’Hare P. 2001 October. Nuclear Translocation and Activation of the Transcription factor NFAT Is Blocked by Herpres Simplex Virus Infection. Journal of Virology 75(20) 9955-9965.
Amasaki Y., Adachi S., Ishida Y., Iwata M., Arai N., Arai K.,
Miyatake S. 2002 July 12. A Constitutively Nuclear Form of NFATx Shows Efficient
Transactivation Activity and Induces Differentiation of CD4+CD8+ T cells. The
Journal of Biological Chemistry 227:28 25640-25648.
Horsley V., Pavlath G. K. 2002 March 5. NFAT: ubiquitous regulator of cell differentiation and adaptation. The Journal of Cell Biology: 156(5) 771-774.
Chen L, Mark Glover J. N., Hogan P. G., Rao A., Harrison S. C. 1998 March 5. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature 392: 42-48
Macian F., Carmen G-R., Rao A. 2000. Gene expression elicited by NFAT in the presence or absence f cooperative recruitment of Fos and Jun. The EMBO Journal 19(17): 4783-4795.
Gomez J., Martinez-A C., Gonzalez A., Garcia A., Rebollo A. 1998. The Bcl-2 gene is idfferentiall regulated by IL-2 and IL-4: role of the transcription factor NFAT. Oncogene 17:1235-1243.
Macian F., Garcia-Cozar F., Im S-H., Horton H. F., Byrne M. C., Rao A. 2002 June 14. Transcriptional Mechanisms Underlying Lymphocyte Tolerance. Cell 109: 719-731.
Crabtree G. R., Olson E. N. 2002. NFAT Signaling: Choreographing
the Social Lives of Cells. Cell 109: S67-S79.
Alan Cubre's Immunology Home Page
Davidson College Biology Department
© Copyright 2002 Department of Biology, Davidson College, Davidson, NC 28035
Send comments, questions, and suggestions to: alcubre@davidson.edu