*This web page was produced as an assignment for an undergraduate course at Davidson College*
CD59: A Membrane-bound Complement Regulatory Protein (mCRP)
Figure 1. Explore this interactive chime image of CD59 protein. This image is courtesy of the Protein Data Bank. http://www.rcsb.org/pdb/
Other Names for CD59
P-18
membrane inhibitor of reactive lysis (MIRL)
protectin
homologous restriction factor-20 (HRF20)
H19
What is CD59?
CD59 is a membrane glycoprotein with a molecular weight of 18-20-kD that plays a crucial role in regulation of the complement system. A terminal product of the complement cascade is the membrane attack complex (MAC, C5b-9). MACs include C5b, C6, C7, C8 and multiple C9 molecules. C5 is first cleaved by a convertase molecule releasing C5b. Through subsequent cleavage processes, C6 is able to bind to C5b, and C7 binds to the C5b6 complex. At this stage a hydrophobic site on the C7 molecule is exposed and it can bind to the target cell membranecomplex becomes hydrophobic and spontaneously binds to the target cell membrane. Binding of C8 occurs when the C8beta protein binds to the C5b molecule of the complex. At this stage, the C8 molecule develops a hydrophobic site and inserts into the cell membrane. Therefore the embedded C5b-8 complex is oriented in a ring-shape which creates a leaky pore through which ions can move. The binding of multiple C9 molecules to the C8a-g protein greatly enlarges the diameter of the MAC and ensures lysis of the target cell.
CD59 blocks the formation of the pore forming membrane attack complex (MAC) by preventing development of a hydrophobic region on the C9 molecule, important for insertion into the target cell membrane. Therefore, a single C9 may bind to the C5b-8 complex but it can not be unfolded to allow binding of multiple subsequent C9 molecules. Therefore MAC formation is haulted and lysis of the target cell does not occur (Davies & Lachmann 1993).
Farkas et. al. (2002) found that CD59 not only blocks the insertion of C9 into MAC but prevents ion channel formation by the C5b-8 complex and the small-size MAC C5b-C9 complex formed by attachment of a single C9 molecule. Therefore, no leaky pore can form. CD59 is critically important in reducing the lytic effect of C5b-8 or C5b-9 during the time interval before the target cell is able to remove the MAC complex by shedding, vesiculation or endocytosis (Hansch & Schin 1998; Farkas et. al. 2002)
Although CD59 is an effective inhibitor of MAC-mediated lysis, Meri et. al (1990) found that CD59 does not prevent perforin-mediated lysis of cells. This may be related to the fact that perforin shows only a 17-21% homology to C9 although some structural similarities exist between perforin and the terminal complement products (C7, C8, C9). GPI anchored proteins have not been found to play a role in cell-mediated lysis by cytotoxic or natural killer cells.
Acosta et. al (2000) found that glycation of the CD59 residues lys41 and his44 associated with increased glucose levels in the body can inhibit complement regulatory function of CD59. This inactivation of CD59 leads to increases in MAC-induced growth factor release from endothelial cells. CD59 is inactivated bound in vitro by the monoclonal antibodies YTH53.1, MEM-43, IF5, and H19 which may act at distinct epitopes (Walsh et. al 1993)
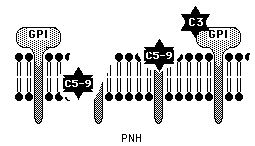
Figure 4. Without important GPI anchored membrane-bound regulatory proteins such as DAF and CD59 complement molecules can bind to the target cell membrane and lyse the cell. This image is used with permission from the University of Virgina Health Science Center. http://hsc.virginia.edu/medicine/clinical/pathology/educ/innes/text/rcd/misc.html
Protein and Gene Structure of CD59 and Relationship to the Ly-6 Protein
The nucleotide sequence for CD59 encodes a polypeptide of 128 amino acids in length. The first 25 polypeptides constitute a signal peptide while the remaining 103 are incorporated in the assembled structure of the protein. Strong disulfide bonding of this tightly folded molecule is made possible through binding of the ten cysteine residues. The binding pattern is as follows:Cys3-26, Cys6-13, Cys19-39, Cys45-63, Cys64-69 (Davies & Lachmann 1993; Petranka et. al 1996). Carbohydrate residues attached to the single N-glycosylation site at Asn 18 may influence the orientation of CD59 on the membrane (Davies & Lachmann 1993).
Human CD59 belongs to a superfamily of molecules characterized by conservation of the arrangement of disulfide bridges but CD59 has a distinct protein sequence from other family members including Ly-6 (Davies & Lachmann 1993; Petranka et. al. 1996).
Similarity between CD59 and the murine glycoprotein Ly-6 has been described extensively in the literature. The protein sequence of CD59 is approximately 25% homologous to Ly-6 but it is unlikely that Ly-6 is the human homologue of CD59 (Petranka et.al; Walsh 1992; Davies & Lachmann 1993). The gene encoding CD59 contains four exons spanning 27 kb. The four exons of CD59 show similarities to the Ly-6 gene (Petranka et.al. 1992). In both proteins the first exon is untranslated and the exon splices occur at similar locations along the DNA (Petranka et.al. 1992; Davies and Lachmann 1993). Further evidence of similarity between the two proteins lies in the fact that the 10 cysteine residues are placed in at similar positions. The proteins are of similar size and are both GPI-anchored.
Contrary to the similarities between CD59 and Ly-6, several striking differences are present. The promoter of CD59 is G+C rich whereas this is not true for Ly-6 (Petranka et. al. 1992). Evidence of differences between CD59 and Ly-6 include the fact that Ly-6 is not present on the surface of murine erythrocytes whereas CD59 is expressed on human erythrocytes. Ly-6 genes are inducible by interferon, and CD59 genes are not (Davies & Lachmann 1993).
The three-dimensional structure of CD59 is similar to that of snake venom toxins including erabutoxin, bungarotoxin, and cardiotoxin (Keiffer et. al 1994; Petranka et. al 1996). These molecules are characterized by a central hydrophobic core with three "finger-like loops, a small fourth loop. CD59 is distinguised by a helical structure in the third loop (Petranka et. al 1996)
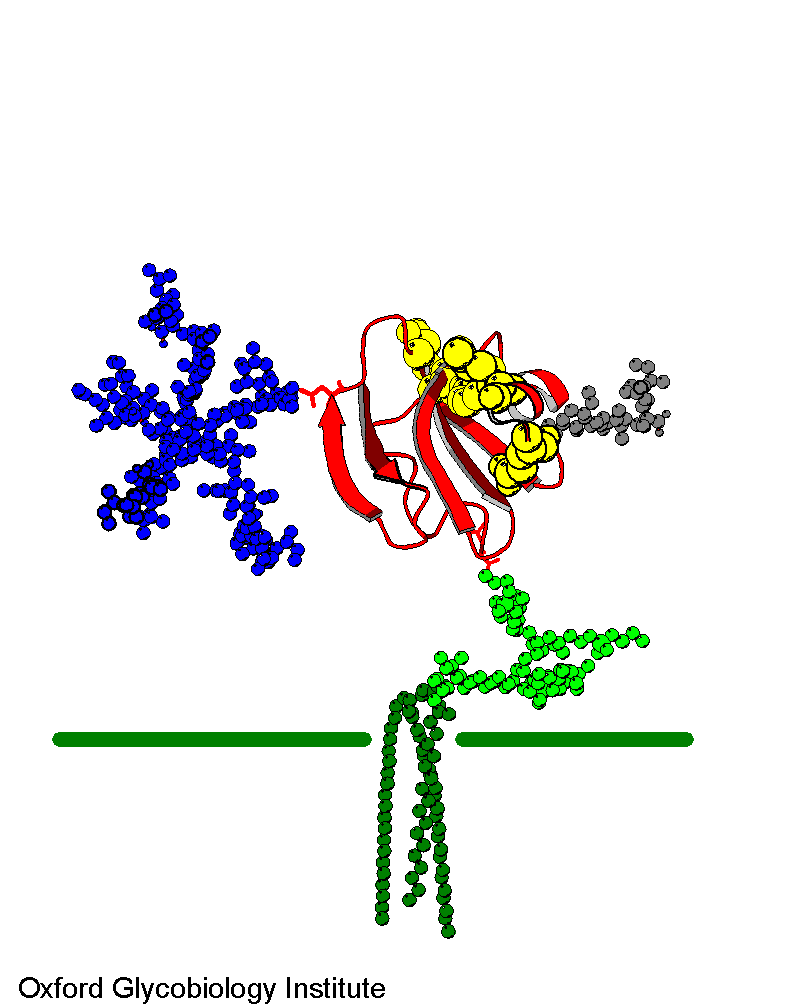
Figure
2. Molecular model of CD59 GPI-anchored in a target cell membrane. There is
a single N-glycosylation site (blue) The active site residues are in yellow.
Image used with permission
from Dr. Mark Wormald of the Oxford Glycobiology Institute.
http://www.bioch.ox.ac.uk/glycob/archive/
Tissue Distribution
CD59 is widely distributed throughout the body to as a regulator of complement-mediated lysis. on both hemopoietic and non-hemopoietic cells (Davies and Lachmann 1993; Walsh et. al 1992). The GPI anchored CD59 protein is present on erythrocytes, leukocytes, fibroblasts and various epithelial surfaces including the kidney, bronchus, pancreas, skin epidermis, and salivary glands (Davies and Lachmann 1993; Walsh et. al 1992). CD59 has also been detected on spermatozoa cells as means of protection from complement-mediated attack by anti-sperm antibodies in the female reproductive tract (Walsh et. al 1992). Little or no CD59 has been detected on B cells or in tissues of the central nervouse system (Davies & Lachmann 1993), The number of CD59 molecules present on each cell has been identified as low as 25,000 and as great as 50,000 depending on the technique used (Davies and Lachmann 1992; Walsh et. al 1992). Initiation of complement attack on a target cell leads to increased expression of CD59 on the cell surface. Soluble CD59 which may be secreted from body cells or GPI-cleaved by phospholipases has been detected in urine, saliva, tears, sweat, amniotic fluid, seminal fluid and breast milk. The function of soluble CD59 is not clear.
Glycophosphoinositol anchoring of membrane proteins and T cell Activation
An important feature of the CD59 molecule is the glycophosphoinositol (GPI) anchor through which is attached to the membrane of body cells. Synthesis of the GPI anchor occurs in the endoplasmic reticulum through the several steps (Jarva & Meri 1999). First an acetylated glucosamine is attached to a phosphoinositol ring. Subsequently the glucosamine is deacetylated, mannose rings are added and ethanolamine is attached (Loertscher & Lavery 2002). The GPI anchor confers certain properties not common in transmembrane anchored proteins. GPI anchored proteins, including CD59, have increased motility along the cell membrane and can be easily released. These properties enable several GPI-linked proteins to transmit activating or inhibiting signals when they are on the surfac of T cells. Upon TcR ligation GPI-anchored proteins cluster with cholesterol and sphingolipids on cell membranes in membrane invaginations called caveolae. Subsequent cross-linking of several GPI-linked proteins (Thy-1, Ly-6, Cd48, and CD59) elevates Ca2+ concentration within the cell and initiates signalling by associating with tyrosine kinase enzymes such as p56lck (Loertscher & Lavery 2002; Jarva & Meri 1999).A study by Loertscher and Lavery (2002) found that GPI-anchored cell surface proteins deliver strong signals supported by the CD28 binding to B7 (Loertscher & Laverty 2002).
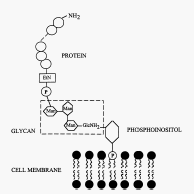
Figure 3. The core structure of glycosylphosphatidyl inositol (GPI) anchor. P, phosphate group; EtN, ethanolamine; Man, mannose; GlcNH2, glucosamine Image used with permission (Jarva & Meri 1999).
http://www.blackwell-synergy.com/links/doi/10.1046/j.1365-3083.1999.00489.x/full/
References
Acosta, J et. al. 2000. Molecular Basis for a link between complement and the vascular complications of diabetes. Proc. Natl. Acad. Sci. USA. 97(10):5450-5.
Bessler, M et. al. 1994. Paroxysmal nocturnal hemoglobinuria (PNH) is caused by somatic mutations in the PIG-A gene. EMBO Journal 13(1):110-117.
Farkas, I et. al. 2002. CD59 blocks not only the insertion of C9 into MAC but inhibits ion channel formation by homologous C5b-8 as well as C5b-9. J. Physiol. 539(2):537-545
Holt, DS et. al. 2001. Targeted Deletion of the CD59 gene causes
spontaneous intravascular hemolysis and hemoglobinuria. Blood 98(2):442-449.
Jarva, H, and Meri, S. 1999. Paroxysmal Nocturnal Hemoglobinuria: the Disease and a Hypothesis for a New Treatment. Scand. J. Immunol. 49:119-125. <http://www.blackwell-synergy.com/links/doi/10.1046/j.1365-3083.1999.00489.x/full/>
Jun-ichi, N et. al. 2002. The Hematopoietic Defect in PNH Is Not Due to Defective Stroma, but Is Due to Defective Progenitor Cells. Blood Cells, Molecules and Diseases 29(2):159-167.
Kieffer, B; Driscoll, PC; Campbell ID et. al. 1994. Three-Dimensional
Solution Structure of the Extracellular Region of the Complement Regulatory
Protein CD59, a New Cell-Surface Protein Domain Related to Snake Venom Neurotoxins
Biochemistry 33: 4471-82.
Loertscher, R, and Laverty P. 2002. The role of glycosyl phosphatidyl insitol (GPI)- anchored cell surface proteins in T-cell activation. Transplant Immunology 9:93-96.
Meri, S et. al. 1990. Human protectin (CD59), an 18-20-kD Homologous Complement Restriction Factor, Does Not Restrict Perforin-mediated Lysis. J. Exp. Med. 172:367-370.
Wormald, M. Oxford Glycobiology Institute. no date. Molecular Model Archives. Accessed 2003 March 14. <http://www.bioch.ox.ac.uk/glycob/>
Petranka et. al. 1992. Structure of the CD59-encoding gene: Further evidence of a relationship to murine lymphocyte antigen Ly- protein. Proc. Natl. Acad. Sci. USA. 89:7876-7879.
Petranka et. al. 1996. Structure-Function Relationships of the Complement Regulatory Protein, CD59. Blood Cells, Molecules, and Diseases 15:281-296.
Protein Data Bank. Structure keyword: 1cdr. Deposited 1994 June 01. Accessed 2003 March 21. <http://www.rcsb.org/pdb/>
Rudd, PM, Morgan, BP, Wormald, MR et. al. The glycosylation of the complement regulatory protein, human erythrocytes CD59. 1997. J. Biol. Chem 272:7229-7244.
University of Virginia, Health Sciences Center. 2000 February 15. Red Cell Disorders. Accessed 2003 March 18. <http://hsc.virginia.edu/medicine/clinical/pathology/educ/innes/text/rcd/misc.html>
Walsh et. al. 1992. The CD59 antigen- a multifunctional molecule.
Tissue Antigens 40:213-220.