Human Interferon gamma (IFN-g)
IFN-g Protein Structure:
As a cytokine, interferon-gamma
is a highly versatile homodimeric protein that plays an essential role
in cell mediated immune responses to viral and mycobacterial infections.
The IFN-g gene is located on chromosome 12 and codes for a 17 kDa protein,
which then undergoes posttranslational glycolysis converting the IFN-g
to a 20-25 kDa glycoprotein1. The molecule that binds to the
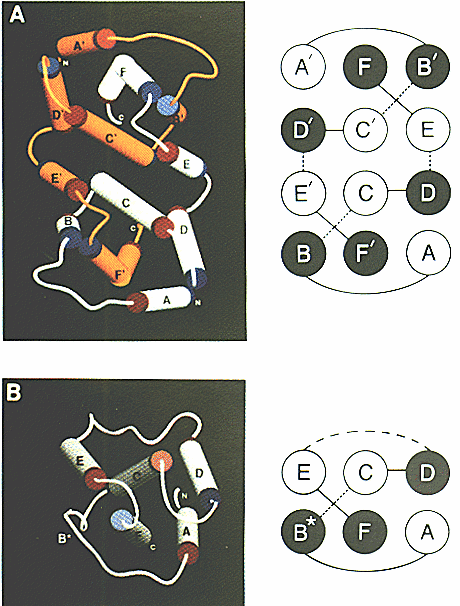
IFN-g receptor is a homodimer as depicted in Fig.1
and has an intricate degree of linkage between the two subunits2.
Each subunit contains six alpha helices and low amounts of beta pleated
sheets. It is a secondary function of IFN-g to decrease the concentration
of intracellular tryptophan, thereby starving intracellular parasites.
Other tryptophan inhibitors have been reported that share the same characteristic
subunit interlocking and high degree of alpha helices found in IFN-g2.
Though there is significant homology of IFN-g between species, the region
between helices A-B and likewise A'-B' is highly variable and may explain
why IFN-g is species specific2.
Figure 1: a) IFN-g homodimer glycoprotein structure
b) IFN-beta protein structure
The cylindrical portions represent alpha helices and are color coded orange
and white
to distinguish between the two separate subunits of the IFN-g homodimer.
This image was borrowed from the following web address pending approval
from its
author Steven Falick.
http://www.cmc.uab.edu/faculty_staff/carson/papers/ifn/ifn_fg4.html
IFN-g signal transduction pathway:
IFN-g receptor, like IFN-g,
is a homodimer with a binding dissociation constant of 10-11
to 10-10 M3. IFN-g receptors are found in high amounts
on the cellular surface of many different cell types such as T cells, B
cells, macrophages, NK cells and fibroblasts1. The receptor
consists of an alpha subunit to which IFN-g binds to and a beta subunit
necessary for signal transduction. As depicted in Fig.2,
the alpha subunit is associated with Janus Kinase 1 (JAK1) and the beta
subunit is associated with Janus Kinase 2 (JAK2). After IFN-g binds to
its receptor the alpha subunit as well as JAK1 and JAK2 are tyrosine phosphorylated.
JAK1 then phosphorylates a tyrosine on signal transducers and activators
of transcription alpha (STAT alpha). After phosphorylation, two STAT alpha
molecules dimerize via interaction between their SH2 domains4.
This dimer, interferon-gamma activator factor (GAF), is able to to enter
the nucleus and bind to interferon-gamma activator sequence. The end result
is an elevated transcription of IFN-g5. Not shown in Fig.2 is
a cofactor protein encoded on chromosome 21 that is necessary for the IFN-g
receptor to function6.
Figure 2: IFN-g signaling pathway resulting in increased transcription
of IFN-g.
The above image was borrowed from the following address pending
approval from its authors.
http://www.grt.kyushu-u.ac.jp/spad/pathway/ifn_gamma.html
IFN-g role in cell mediated immune responses:
IFN-g is produced by NK cells,
dendritic cells, cytotoxic T cells, Progenitor Th0 cells and Th1 cells1.
One of IFN-g's main roles in cell mediated immune responses is its antiviral
activity. IFN-g has no viral specificity and inhibits the spread of viruses
containing either RNA or DNA7. There are several mechanisms
by which IFN-g reacts to viral infection. In conjunction with CD40, IFN-g
binds to and activates macrophages, which are then able to kill intracellular
pathogens such as viruses8. Furthermore, bound IFN-g causes
the macrophage to produce elevated amounts of both MHC class I and II molecules,
thus increasing the macrophage's presentation of foreign peptides. A second
antiviral mechanism of INF-g is to shutdown the replication of viral DNA
or RNA and to rid the cell of pathogen without killing it8.
In addition to its antiviral activity, IFN-g also plays a role in delayed
type hypersensitivity by binding to macrophages and eliciting the release
of inflammatory mediators8.
IFN-g role in immunoregulation:
In accordance with its
elevated syntheses in response to mycobacterial and viral infections, IFN-g
directs several immunoregulatory mechanisms that enhance the cytokine's
function. As previously mentioned, IFN-g increases the production of MHC
class I and II, which in turn increase the likelihood that the infected
cell will be recognized as such. To further facilitate the loading of peptide
onto MHC, IFN-g also upregulates the transcription of HLA-DM, tapasin and
TAP genes8. IFN-g also influences cell differentiation of the
progenitor Th0. By increasing Th1 differentiation from Th0 progenitor cells,
IFN-g in turn inhibits differentiation into Th2 cells9. Isotype
switching in B cells to IgG is also enhanced by IFN-g, presumably because
IgG activates the compliment system and apsonizes extracellular pathogens
resulting in their uptake by phagocytic cells8,10.
Effects of IFN-g deficiencies and miscellaneous information:
For proteins with such
diverse and profound effects as IFN-g it is not only important that it
be synthesized when needed, it is equally important that IFN-g not linger
around when its presence is unnecessary. There are two ways of preventing
IFN-g from building up after its activating signal has been removed, both
of which are self regulatory. Along IFN-g's mRNA 3' untranslated end is
a sequence (AUUUA)n, that in turn reduces the mRNA's half-life.
Secondly, activated T cells in addition to transcribing IFN-g also transcribe
another protein that prompts cytokine mRNA destruction8. As
a result, once the activating signal for increased IFN-g production is
removed the levels of IFN-g quickly return to normal. Not surprisingly,
a deficiency or mutant form of IFN-g has a wide range of effects including
an elevated risk to viral and bacterial infections.
References:
1Gamma-Interferon. <http://www.anticancer.net/resan/gamma_interferon.html>
Accessed 2000 Feb 19.
2Falick SE.1991 Feb. Three-dimensional structure of recombinant human interferon-gamma. <http://www.cmc.uab.edu/faculty_staff/carson/papers/ifn/ifn.html> Accessed 2000 Feb 19.
3Aguet M, Dembic Z, Merlin G. 1988. Molecular cloning and expression of the human interferon-gamma receptor. Cell 55:273-280.
4Shuai Ke, Horvath CM, Tsai Hung LH, Qureshi SA, Cowburn
D, Darnell JE. 1994 Interferon activation of the transcription factor Stat91
involves
dimerization through SH2-phosphotyrosyl peptide
interactions. Cell 76:821-828.
5Signaling pathway mediated by interferon-gamma. <http://www.grt.kyushu-u.ac.jp/spad/pathway/ifn_gamma.html> Accessed 2000 Feb 19.
6Hemmi S, Bohni R, Stark G, Di Marco F, Aguet M. 1994. A
novel member of the interferon receptor family complements functionalityof
the murine
interferon-gamma receptor in human cells. Cell 76:803-810.
7Finter NB. 1966. Interferons. Amsterdam: North-Holland Publishing Company. p 4-12.
8Janeway CA, Travers PT, Walport M, Capra JD.1999. Immunobiology:
The immune system in health and disease 4th ed. Union Square
West, New
York, NY: Elsevier Science Ltd/Garland Publishing.
p 136, 263, 288, 301-301, 483.
9Mosselaar JJ. 2000. Anti-interferon-gamma, clone 45-14.
Product information INF-gamma. <http://www.iqproducts.nl/pfcinfg.htm>
Accessed
2000 Feb 19.
10Campbell NA. 1994. Biology. New York, NY: The Benjamin/Cummings
Publishing Company, Inc. p 864.