This web page was produced as an assignment for an undergraduate course at Davidson College.
Figure 1. NFAT and AP-1 (Fos and Jun) bound to DNA. NFAT and AP-1 DNA-binding regions form a tight complex with each other and with DNA. The yellow structure represents NFAT, the two tight pink coils represent Fos and Jun, and the purple double helix represents DNA. (RCSB Protein Data Bank, 1A02)
What is NFAT?
NFAT = nuclear factors of activated T cells
NFAT is a family of transcription activators especially known for its role in activating the transcription of cytokine genes in activated T cells. The NFAT family is evolutionarily related to the Rel/NFκB family (Hogan et al., 2003). NFAT was originally identified in the promoter system of the cytokine interleukin-2 (IL-2), but is also known to regulate other promoter regions such as those of cytokines IL-3, IL-4, and tumor necrosis factorα (TNFα). Thus, NFAT plays a significant role in T-cell activation. Within the immune system, the NFAT proteins are expressed in T cells, B cells, mast cells, and natural killer (NK) cells (Lyakh et al., 1996), yet its function is most defined within T cells. As well as regulating the transcription of genes involved in the immune system, NFAT plays important roles in other bodily systems. While this page is dedicated to the immunological roles of the NFAT proteins, it does not disregard its importance in other vertebrate systems such as the vascular system and neural system (Nuclear Factor of Activated T cells, Cytoplasmic, Calcineurin-Dependent 3; NFATC3). NFAT's activity is defined by its translocation from its inactive form in the cytoplasm to its active form in the nucleus, where it can bind to promoter regions on cytokine genes and activate their transcription. Its activity is aided by calcineurin, a Ca2+/calmodulin-dependent phosphoserine/threonine phosphatase, which plays a key role in NFAT's ability to translocate into the nucleus and act as a transcription factor (Hogan et al., 2003).
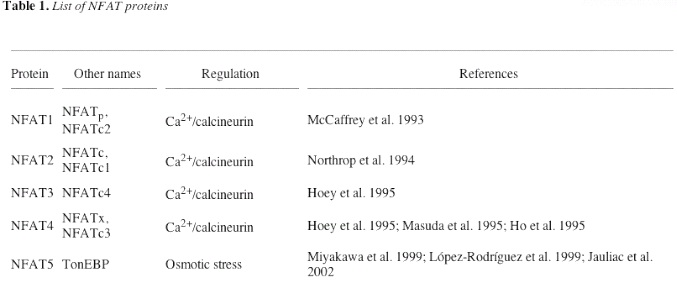
There are currently five known members of the NFAT family as shown in the table below (Hogan et al., 2003). NFAT5 stands out because it does not follow the same calcium/calcineurin signaling pathway that the other four other NFAT proteins exhibit. The NFAT members that are focused on when looking at NFAT's roles in the immune system are the first four NFAT members: NFAT1, NFAT2, NFAT3, and NFAT4. The NFAT proteins that will be referred to in this page are these four NFAT members.

Table 1. List of NFAT proteins. Includes other names for each NFAT protein and the controllers of their regulation (Hogan et al., 2003).
Protein structure defines NFAT signaling
NFAT is either found as a monomer in solution or bound to DNA. As a monomer it is found in the cytoplasm of the cell in which it is expressed. As a unit bound to DNA, it is found in the nucleus (Masuda et al., 1995). The four NFAT (NFAT1-NFAT4) genes encode proteins that range from approximately 700-1,100 amino acid residues. NFAT amino acid structure is divided into four large domains and many subdomains within these domains. Each domain is characterized by a different function in the NFAT signaling pathway. The four domains are: the N-terminal domain, the NFAT homology region, the Rel homology domain, and the C-terminal domain. (Masuda et al., 1997; Park et al., 2000 ; Zhu and Mckeon, 1999).
Figure 2. The NFAT domains. The regulatory domain is where calcineurin binding and dephosphorylation/phosphorylation occur. The red circles indicate phosphate groups that can be removed by calcineurin and the black circle on the SRR-2 domain indicates a phosphate group that can not be dephosphorylated by calcineurin. The A and B motifs indicate the calcineurin binding sites CnBP-A and CnBP-B (Hogan et al., 2003).
The NFAT homology region (NFAT-h) is a domain within the larger regulatory domain. It is made up of about 300 amino acid residues on the N-terminal side of the NFAT protein. NFAT-h is a serine-rich region that is highly conserved among the four NFAT proteins. Within this region, there are nine conserved motifs that are directly involved in different aspects of the NFAT signaling pathway (Park et al., 2000). These motifs contain sequences that either serve as phosphorylation/dephosphorylation sites, calcineurin/Crm-1 binding sites, or translocation directors. Many of these motifs are known to have direct functions that ultimately aid in the translocating of NFAT into the nucleus (Park et al., 2000).
The goal of cytokine transcription must be preceded by the translocation of NFAT into the nucleus. The clustering and activation of T-cell receptors on the cell surface and the actions that follow this activation must precede NFAT translocation. T-cell receptor activation initiates a signal transduction pathway that leads to an influx of Ca2+ into the cytoplasm from channels in the ER membrane and from Ca2+ release activated Ca2+ channels (Crac channels) in the cell membrane (Crabtree and Olson, 2002). The Ca2+/calmodulin complex binds and activates calcineurin by triggering a conformational change in calcineurin. This conformational change activates calcineurin's phosphatase activity and enhances calcineurin's affinity for NFAT (Zhu and Mckeon, 1999). Before dephosphorylating NFAT, active calcineurin binds NFAT at two of the nine conserved motifs within the NFAT-h region. It binds calcineurin-binding peptide A (CnBP-A) and calcineurin-binding peptide B (CnBP-B) (Park et al., 2000). Arambaru et al. (1998) mapped CnBP-A in NFAT1 as the sequence SPRIEITPS. The consensus sequence of CnBP-A for all of the NFAT proteins is PxIxIT (Hogan et al., 2003) CnBP-A lies in the N-terminal end of the NFAT-h region. Park et al. (2000) found that there was in fact another sequence that plays as significant of a role as CnBP-A in binding calcineurin. This sequence, CnBP-B, is found in all four NFAT isoforms and is located in the C-terminal end of the NFAT-h region. CnBP-A and CnBP-B interact with distinct and nonoverlapping sites on calcineurin and the strength of interaction at each site differs with each isoform of NFAT (Park et al., 2000).
Once bound to CNBP-A and CNBP-B on NFAT, calcineurin dephosphorylates NFAT, removing the phosphates from the serine-rich motifs on the NFAT-h region. These serine-rich motifs are the serine-rich gatekeeper region (SRR-1) and the three SPxx repeat motifs (SP-1, SP-2 and SP-3). They are hyperphosphorylated in the cytoplasm before calcineurin exhibits its phosphatase activity. Dephosphorylation of these NFAT-h motifs exposes the nuclear localization signal (NLS) motif causing the nuclear import receptor importin-α to bind to the NLS. This leads to NFAT's immediate import into the nucleus (Crabtree and Olson, 2002). Calcineurin remains bound to NFAT at NFAT's nuclear export signal (NES) subdomains and and is co-transported with NFAT into the nucleus. There are two NES subdomains (NES1 and NES2) within the NFAT-h region (Zhu and Mckeon, 1999). By binding to NFAT at its nuclear export signal sequences, calcineurin masks this signal sequence, thus preventing NFAT's export out of the nucleus. In the nucleus, calcineurin competes with Crm-1, a receptor for nuclear export signal, for the NES binding site. When a cell no longer has a constant influx of Ca2+ through Ca2+ release activated channels, Crm-1 binds to NFAT's NES leading to nuclear export. Yet, in a cell that is presented with an influx of Ca2+, calcineurin and Crm-1 compete for the NES binding site, but calcineurin wins because it binds NFAT with a higher affinity than does Crm-1 (Zhu and Mckeon, 1999). Not only does calcineurin's presence in the nucleus aid in maintaining NFAT within the nucleus, its presence is required for NFAT to form effective NFAT-transcriptional complexes (Zhu and Mckeon, 1999).
The Rel homology domain is NFAT's DNA-binding domain. It is another domain that is highly homologous among the NFAT proteins. This domain is designated the Rel homology domain because of its resemblance to the Rel homology region of Rel-family transcription factors. NFAT binds DNA and another transcription factor AP-1, a Fos-Jun dimer, via its Rel homology domain (Figure 1). AP-1 and NFAT require cooperative association with each other in order for either one to be able to bind DNA and activate transcription. NFAT has a tight yet discontinuous interaction with AP-1. NFAT's N-terminal domain has large contact surface with the single α helices Fos and Jun while its C-terminal has very small contact with Fos. The tight NFAT:AP-1 complex forms a tight complex with the promoter region of the IL-2 gene and other cytokines genes (Chen et al., 1998). This binding leads to transactivation of cytokine genes via NFAT's N-terminal and C-terminal transactivation domains. The N-terminal transactivation domain is located at the end of the N terminal (Aramburu et al., 1998). The C-terminal transactivation domain is within the C-terminal region and exhibits very little homology among the NFAT family members (Masuda et al., 1997).
Once transcription of IL-2 and other cytokine genes has been induced, the Ca2+ signal subsides causing calcineurin to dissociate from NFAT. The kinases protein kinase A (PKA) and GSK-3β are responsible for phosphorylating NFAT. Phosphates are added to the serine-rich regions of the NFAT-h region resulting in the shielding of the NLS (Park et al., 2000). Calcineurin's dissociation from NFAT opens up the NES for binding by Crm-1. The shielding of the NLS due to phosphorylation along with the exposure of the NES and subsequent binding by Crm-1 directs NFAT out of the nucleus and into the cytoplasm. Yet, unlike calcineurin, Crm-1 does not co-transport with NFAT during NFAT's translocation and therefore is not bound to NFAT in the cytoplasm. The phosphorylated form of NFAT remains inactive in the cytoplasm until the next influx of Ca2+ into the cytoplasm.
Figure 3. NFAT and AP-1 signaling pathways. The pathway begins with the activation of T-cell receptors. T-cell receptor activation initiates a signal transduction pathway that leads to an influx of Ca2+ into the cytoplasm. Ca2+ activates calcineurin causing calcineurin to dephosphorylate NFAT. Once dephosphorylated, NFAT can translocate into the nucleus and activate transcription along with the AP-1 transcription factor. T-cell receptor activation also initiates the MAP kinase pathway (left side of figure) that leads to the activation of the Fos and Jun α helixes that make up AP-1 (www.callisto.si.usherb.ca/ ~bcm514/3a.html).
Inhibitors of NFAT signaling
Many of the known inhibitors of NFAT signaling inhibit NFAT indirectly by affecting calcineurin's activity.
Drugs that inhibit NFAT signaling:
The two primary drug inhibitors of calcineurin are cyclosporin-A (CsA) and FK506. Each of these drugs bind a type of protein called an immunophilin. An immunophilin is an intracelluar immunosuppressant-binding protein that forms a complex with the immunosuppressant it binds. CsA forms an immunophilin-immunosuppressant complex with the immunophilin cyclophilin. FK506 binds and forms a complex with FK506-binding proteins (FKBPs). Cyclophilins and FKBPs are ubiquitous proteins increasing the likelihood that immunophilin-immunosuppressant complexes will form if a cell is exposed to CsA or FK506. The immunophilin-immunosuppressant complexes cyclophilin-CsA and FKBP-FK506 bind calcineurin and inhibit its ability to bind and dephosphorylate NFAT. CsA and FK506 are the primary immunospressants used in order to prevent graft rejection after an organ transplant because of their ability to prevent NFAT from being translocated into the nucleus and ultimately leading to an immune response (Breuder et al., 1994).
Cellular inhibitors of NFAT signaling:
There are four known cellular inhibitors of calcineurin phosphatase complexes. One is AKAP79, a scaffold protein that was the first found inhibitor of calcineurin. AKAP79 binds calcineurin and prevents its access to substrates. Another is the CAIN protein which directly blocks calcineurin activity. Deletion of the gene that encodes CAIN leads to T cell hyperactivation. This indicates that CAIN plays a role in regulating calcineurin. The Calcineurin B homolog, CHP, inhibits calcineurin by binding to CnA, a calcineurin subunit, without inducing its activation (Crabtree and Olson, 2002).
Viruses that inhibit NFAT signaling:
Calcineurin is specifically targeted by viruses such as the African Swine fever, Trichoderma, Streptomyces, and leukemia viruses. The African Swine fever virus produces the A238L protein. A238L acts similarly to the drugs CsA and FK506 in that it binds calcineurin and prevents the translocation of NFAT into the nucleus (Crabtree and Olson, 2002).
Result of NFAT mutation
The significance of NFAT in T-cell activation and other immune functions is made very apparent when the genes encoding NFAT proteins are either deleted or mutated enough to significantly alter NFAT's function as a transcription factor. In most cases, huge effects are not seen unless more than one NFAT gene is removed because the proteins in the NFAT family have relatively redundant functions. Both NFAT1 and NFAT2 need to be deleted in order to significantly reduce cytokine production in T cells. The deletion of both NFAT1 and NFAT4 produces a large bias toward TH2 cytokine production in mice (Hogan et al., 2003). Some exceptions to this rule are seen in the isolated deletions of NFAT2, NFAT4 and NFAT1. The deletion of NFAT2 leads to defects in thymic development and a reduction in the activation of IL-2, IL-4, and other cytokines. NFAT4 deletion also results in defects in thymic development. The absence of NFAT4 leads to a loss of double positive cells through programmed cell death (Crabtree and Olson, 2002). NFAT1 deletion results in the reduction of cytokine production in mast cells (Hogan et al., 2003), immune hyperactivation, and allergic responses (Crabtree and Olson, 2002).
References
Aramburu, J., Garcia-Cozar, F., Raghavan, A., Okamura, H., Rao, A., Hogan, P.G. Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Molec. Cell 1: 627-637, 1998.
Breuder, T., Hemenway, C.S., Movva, N.R., Cardenas, M., and Heitman, J. Calcineurin is essential in cyclosporin-A- and FK506-sensitive yeast strains. Proc. Natl. Acad. Sci. USA 91: 5372-5376, 1994.
Chen, L., Glover, J.N., Hogan, P.G., Rao, A., and Harrison, S.C. Structure of the DNA-binding Domains from NFAT, Fos and Jun bound specifically to DNA. Nature 392: 42-48, 1998.
Crabtree, G.R. and Olson, E.N. NFAT signaling: choreographing the social lives of cells. Cell 109: S67-S69, 2002.
Hogan, P.G., Chen, L., Nardone, J., and Rao, A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes & Development 17: 2205-2232, 2003.
Lyakh, L., Ghosh, P., and Rice, N.R. Expression of NFAT-family proteins in normal human T cells. Molec. Cell Biol. 17: 2475-2484, 1996.
Masuda, E.S., Liu, J., Imamura, R., Imai, S., Arai, K., and Arai, N. Control of NFATx nuclear translocation by a calcineurin-regulated inhibitory domain. Mol. Cell Biol. 17: 2066-2075, 1997.
Masuda, E.S., Naito, Y., Tokumitsu, H., Campbell, D., Saito, F., Hannum, C., Arai, K., and Arai, N. NFATx, a novel member of the nuclear factor of activated T cells family that is expressed predominantly in the thymus. Molec. Cell Biol. 15: 2697-2706, 1995.
"Nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 3; NFATC3." 14 Sep 2005. Online Mendelian Inheritance in Man. 14 Mar 2006 <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=omim>.
Park, S., Uesugi, M., and Verdine, G.L. A second calcineurin binding site on the NFAT regulatory domain. Proc. Natl. Acad. Sci. USA 97: 7130-7135, 2000.
Zhu, J. and Mckeon, F. NF-AT activation requires suppression of Crm1-dependent export by calcineurin. Nature 398: 256-260, 1999.
© Copyright 2006 Department of Biology, Davidson College, Davidson,
NC 28036
Send comments, questions, and suggestions to: kaacker@davidson.edu