This web page was produced as an assignment for an undergraduate course at Davidson College.
My Favorite Immunology Protein: Caspase Activated Deoxyribonuclease (CAD)
In literature, this protein has been alternatively referred to as: Caspase Activated Nuclease (CPAN), DNA Fragmentation Factor 40 (DFF-40), DFF2, DFFB, and other variations on these combinations.
Introduction
Human Caspase Activated Deoxyribonuclease (CAD) is the protein product of a single gene with 3034 nucleotides located on chromosome 1 at the position 1p36.3. It is expressed in various tissues and cell types including leukocytes, the spleen, thymus, kidney, heart, pancreas, ovary, colon, prostate, and placenta (Peri S, et al., 2006).
CAD is an ultimate endonuclease in the apoptotic pathway initiated by Fas and Fas-L. When activated by caspase-3, CAD is responsible for cleaving DNA into the characteristic ~200 bp fragments of apoptotic cells.
Structure
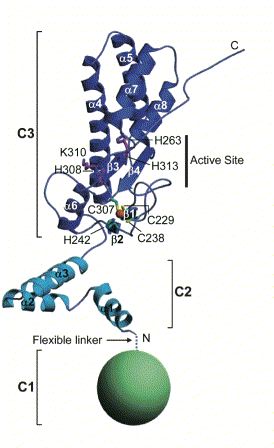
Human CAD is made up of 338 amino acids and has an approximate weight of 40 kD. CAD contains three domains described by Woo EJ, et al. (2004).The first domain (N-terminal CAD domain or C1) consists of amino acids 1-85 and remains to be fully characterized. The second domain (C2) consists of amino acids 86-131 that make up three separate a chains. Finally, the third, largest, and functionally most important domain (C3) contains amino acids 132-328. These amino acids fold to form five a helices, four b sheets, and a loop at the C-terminal. Together C1, C2, and C3 make up a CAD monomer (Fig 1).

Figure 1. Cartoon Structure of CAD monomer. Note the three key domains of CAD: C1 (green sphere), C2 (light blue), C3 (dark blue). Important amino acids, secondary structure, regions (i.e. active site), and termini have all been labeled. Image courtesy of: (Woo EJ, et al., 2004), permission pending.
In order to function in vivo, two CAD monomers come together to form a homodimer with vertical symmetry (Fig 2). The two C3 domains, in particular, associate with each other such that they form a cavity inside which a section of double-stranded DNA can fit (Woo EJ, et al., 2004). Several residues in and around the a4 helix of CAD (beginning at amino acid 156) are essential for the binding the major groove of DNA to the cavity (Reh S, et al., 2005). In fact, CAD's ability to bind DNA is disrupted with a single amino acid change, when lysine at position 155 is replaced with glycine (Reh S, et al., 2005, citing Korn C, et al., 2005).
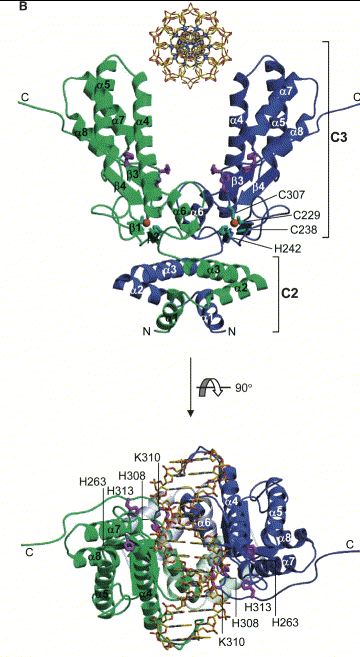
Figure 2. Cartoon Structure of the CAD homodimer with an associated section of DNA. One CAD monomer appears in green while the other appears in blue. The CAD/DNA complex has been rotated in the bottom portion of the image for alternative viewing. Key amino acids, secondary structure, domains, and termini have been labeled. Image courtesy of: (Woo EJ, et al., 2004), permission pending.
The active site of CAD (shown in Fig 1) allows CAD to bind the minor groove of a strand of DNA (Reh S, et al., 2005) and controls the catalytic ability of the protein. The highly conserved histidine residues 242, 263, 313, 308 present in the active site are essential for maintaining the nuclease function of CAD (Meiss G, et al., 2001). Furthermore, magnesium and zinc in moderate concentrations must be associated with CAD to preserve function (Woo EJ, et al., 2004).
You can explore the structure of CAD, especially the portions mentioned above, using the interactive JMOL image below (Fig 3).
Figure 3. JMOL image of CAD. Image courtesy of the Protein Data Bank.
Function
CAD is an final player in the extrinsic pathway of programmed cell death typically initiated by Fas/Fas-L (Fig 4). When a Fas ligand binds to a Fas receptor on another cell, conformational changes occurs in Fas such that its intracellular portions are able to associate with other proteins such as FADD and a caspase cascade is initiated. In particular, caspase-8 becomes activated and activates capsase-3 in turn. In its typical inactive state, CAD resides in the cytosol of a cell and is associated with its inhibitor, ICAD-L. However, activated caspase-3 is able to cleave ICAD-L and thus activate CAD. Activated CAD can translocate into the nucleus of a cell via a nuclear localization signal (Nagata S, 2004) and begin to cleave chromosomal DNA.
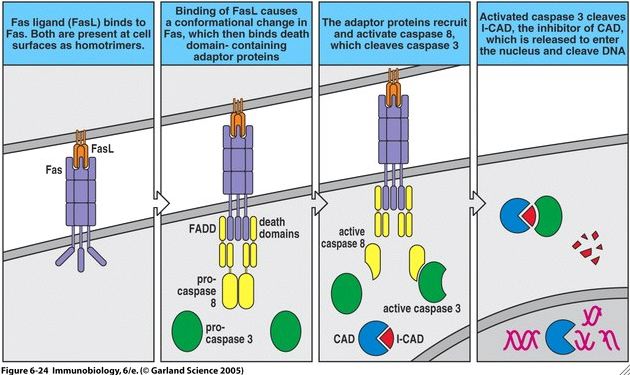
Figure 4. Fas/Fas-L mediated apoptotic pathway. Note that CAD appears in light blue, ICAD-L appears as red, and caspase-3 appears as green. Image courtesy of: (Janeway CA, et al., 2005), permission pending.
CAD functions at a neutral pH and is classified as an endonuclease because it cleaves DNA at the internal portions of a strand, rather than from the ends (Nagata S, 2004). Furthermore, CAD's nuclease activity is specific: it generally cuts in A/T rich regions of DNA and it only cleaves double stranded DNA (Nagase S, et al., 2003). CAD fragments DNA in ~200 bp units because nucleosomes (~200 bp of wound DNA) are too large to fit in the cavity of CAD. Thus, only unwound portions of DNA (i.e. the portions between nucleosomes) are cleaved by CAD. For a visualization of nucleosomes please see Dr. Campbell's Quick Lesson on Apoptosis Production of 200 bp DNA Fragments.
ICAD-L acts as both an inhibitor and chaperone of CAD. On one hand, ICAD-L must be cleaved from CAD to activate it. On the other hand, CAD requires an association with ICAD-L to fold properly. Indeed, ICAD-L knockout mice cannot produce functional CAD (Nagata S, 2004). Thus, there exists some necessary internal control of the CAD molecule because it cannot be expressed properly without the co-expression of its inhibitor, ICAD. CAD has the potential to wreak havoc on a cell if it is not properly in check by this mechanism.
Apoptosis is essential for the clearance of pathogen-infected cells, cancerous cells, lymphocytes that do not pass positive and/or negative selection, and other cells during development. However, immunologists have not determined whether DNA fragmentation that results from CAD activity is essential for programmed cell death. Indeed, caspases activated in the caspase cascade cleave other key factors in the cell that are necessary for survival and normal processes, including ribosomal proteins and RNA polymerase (Nagata S, 2004). Whether CAD DNA fragmentation is a necessary part of apoptosis or simply a result of apoptosis remains to be elucidated.
Knockouts, Mutations, and Associated Pathology
CAD knockout mice (Nagase H, et al., 2003) and fruit flies (Nagata S, 2003, citing Mukae N, et al., 2002) have been successfully bred. These animals are generally healthy, but logically, the numbers of cells in these animals that exhibit DNA fragmentation after certain extrinsic apoptotic initiation signals are drastically reduced. The DNA fragmentation that occurs in some cells of these animals may be the result of compensation by another nuclease protein called DNase II (Nagata S, 2003).
Interestingly, cancerous cells often contain a mutation their CAD gene in addition to other aberrations. In fact, Hsieh SY, et al. (2005) have shown a certain recurrent homologous recombination/deletion in the CAD gene in 13 out of 20 human heptoma samples. Because cancer cells divide uncontrollably, it is understandable that their genes that encode proteins involved in apoptotic pathways are mutated. Mutations in CAD may be just one of the may genes that becomes mutated in the progression of certain cancers.
Finally, Uegaki K, et al. (2005) have shown that the C1 domain of CAD "aggregates to form amyloid fibrils, without a lag time, under the conditions of low pH and the presence of anions." Thus, if C1 is perhaps mistakenly cleaved from CAD in large quantities, amyloid fibrils could form and diseases like Alzheimer's could result. Indeed, improper processing of CAD may play a role in the initiation or progression of Alzheimer's disease.
Drug Binding
No drugs are listed as binding to CAD on OMIM. However, since moderate doses of zinc are required for the proper function of CAD, the incubation of the Zn2+ chelator, o-phenantroline, effectively inactivates CAD (Woo EJ, et al., 2004). Furthermore, if CAD is treated with too much ZnCl2 solution, CAD DNase activity is disrupted. Thus, drugs that contain metal chelators and or zinc ions may affect the function of CAD in humans.
For more information about CAD, please visit:
The Human Protein Reference Database
References
Hsieh SY, Chen WY, Yeh TS, Sheen IS, Huang SF. 2005 Sept 29. High-frequency Alu-mediated genomic recombination/deletion within the caspase-activated gene in human heptoma. Oncogene 24: 6584-9.
Janeway, CA, Travers P, Walport M, Shlomchik MJ. 2005. Immunobiology: The immune system in health and disease, 6th edition. New York: Garland Publishing.
Meiss G, Scholz SR, Korn C, Gimadutdinow, Pingoud A. 2001. Identification of functionally relevant histidine residues in the apoptotic nuclease CAD. Nuc Acid Res 29: 3901-3909.
Nagase H, Fukuyama H, Tanaka M, Kawane K, Nagata S. 2003. Mutually regulated expression of caspase-activated DNase and its inhibitor for apoptotic DNA fragmentation. Cell Death and Diff 10: 142-143.
Nagata S, Nagase H, Kawane K, Mukae N, Fukuyama H. 2003. Degradation of chromosomal DNA during apoptosis. Cell Death and Diff 10: 108-116.
Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK,Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, Ibarrola N, Deshpande N, Shanker K, Shivashankar HN, Rashmi BP, Ramya MA, Zhao Z, Chandrika KN, Padma N, Harsha HC, Yatish AJ, Kavitha MP, Menezes M, Choudhury DR, Suresh S, Ghosh N, Saravana R, Chandran S, Krishna S, Joy M, Anand SK, Madavan V, Joseph A, Wong GW, Schiemann WP, Constantinescu SN, Huang L, Khosravi-Far R, Steen H, Tewari M, Ghaffari S, Blobe GC, Dang CV, Garcia JG, Pevsner J, Jensen ON, Roepstorff P, Deshpande KS, Chinnaiyan AM, Hamosh A, Chakravarti A, Pandey A. 2006. The Human Protein Reference Database. <http://www.hprd.org>. Accessed 2006 Mar 17.
Reh S, Korn C, Gimadutdinow O, Meiss G. 2005 Dec 16. Structural basis for stable DNA complex formation by the caspase-activated DNase. J of Bio Chem 280: 41707-41715.
Shigekazu N. 2005. DNA degradation in development and programmed cell death. Annu Rev Immunol: 853-875.
Uegaki K, Nakamura T, Yamamoto H, Kobayashi A, Odahara T, Harata K, Hagihara Y, Ueyama N, Yamazaki T, Yumoto N. 2005. Amyloid fibril formation by the CAD domain of caspase-activated DNase. Biopolymers 79: 39-47.
Woo EJ, Kim YG, Kim MS, Han WD, Shin S, Robinson H, Park SY, Oh BH. 2004 May 21. Structural mechanism for inactivation and activation of CAD/DFF40 in the apoptotic pathway. Molecular Cell 14: 531-539.
Return to Jackie's Immunology Main Page