Analysis of PFK cDNA sequence
Phosphofructokinase is an enzyme involved in the glycolysis pathway. The following is some interesting material developed using the MacDNAsis program.

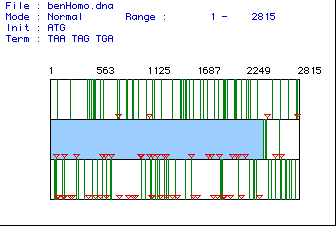
Figure 1. Any given DNA sequence can be translated using 3 different reading frames, depending on which base you start with. This shows the three possible reading frames for the cDNA sequence, with the middle (hilighted) being the actual reading frame, with the ORF running from bp 77 to 2419 bp. The red triangles mark the start codons, and the green lines show stop codons. The center reading frame is the correct one because it has the greatest amount of space from a start codon to a stop codon.
A translation of the cDNA sequence for Homo sapiens PFK came up with a predicted molecular weight of 85178.08 D.
Figure 2. A Kyte and Doolittle hyropathy plot shows a possible 9 membrane spanning domains (portions above the 1.8 threshold). This program is used to predict where membrane spanning domains might occur based on the hydrophobicity of any given segment (in this case, the predicted hydrophobicity of every 8 aa s). Judging by this model, with 9 possible membrane spanning domains predicted, this makes PFK a possible integral membrane protein.
Figure 3. An antigenicity plot shows those domains of DNA which are most hydrophilic, and thus most exposed and available for epitope binding. Other factors are considered in the prediction as well, such as aromaticity of aa residues. A monocolonal antibody to the section from a.a.s 90 to 100 would be a good candidate for successful binding, because it is the longest very hydrophilic region.

Figure 4. The (partial) predicted secondary structure
for a portion of the cDNA sequence. Compare this to the rasmol
image of PFK (MMDB Id: 2854 PDB Id: 2PFK).
Figure 5. Amino acid sequence alignment for the translated PFK sequence with other organisms' sequences. This shows the degree of homology between homo sapiens PFK and the other 4 genome project organisms. benHomo is the Homo sapiens sequence translated by MacDNAsis, BenDroso is a sequence for Drosophila melanogaster, benelegant is C. elegans, benhomo a Homo sapiens sequence, BenMusMus is mouse, and benyeast is S. cerevisiae.
Figure 6. This shows the degree of similarity of the various aa sequences to the Homo sapiens sequence. The species are the same as in figure 5. The higher the percent similarity between sequences, the closer the species are on the philogenetic tree of life, in theory, and thus more homology.
Direct correspondence to: bebuxton@davidson.edu