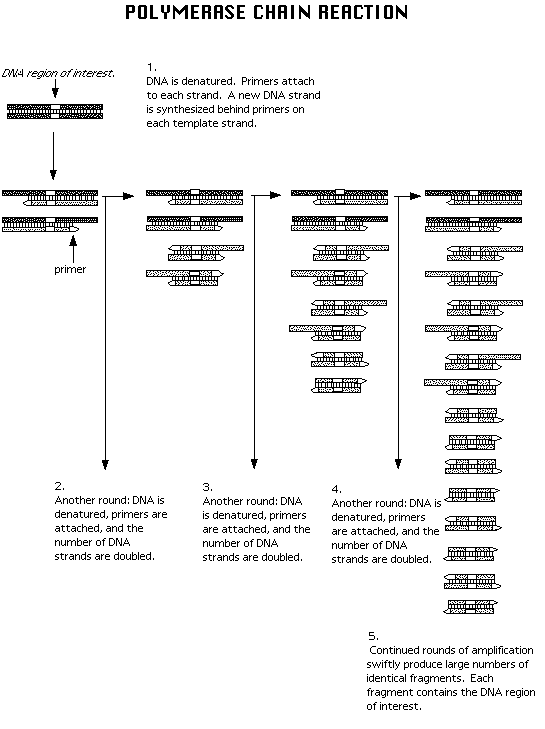
The polymerase chain reaction, or PCR, is a method that allows production of billions of copies of a specific DNA segment in just hours. PCR was first described in 1985, but it was inspired by a bacterium that is millions of years old.1, 3Thermus aquaticus, a bacterium residing in the hot springs of Yellowstone National Park, relies upon an enzyme that functions in the extreme temperatures of the springs.3 PCR has a broad range of uses, including, but not limited to, amplification of DNA isolated from extinct organisms, crime scene analysis, and disease detection. DNA must first be isolated and purified in order to run an uninhibited PCR reaction. One intact strand of the DNA region of interest must be present within the sample. DNA must be thoroughly washed in order to limit the possibility that the PCR will be inhibited by impurities in the sample.
After DNA is isolated, three main steps are required in the PCR process.
1) Double-stranded DNA (dsDNA) is heated to 950 C for 30 seconds up to a few minutes, thus separated into its single stranded components.
2) The single-stranded DNA (ssDNA) is allowed to cool to 550 C, so one primer can hydrogen bond to each existing ssDNA template.
3) Taq DNA polymerase, isolated from Thermus aquaticus, extends the primers by adding nucleotides complementary to the template strand. Usually, Taq is most effective at 750 C, the temperature in the hot springs from which the enzyme was isolated.
Steps 1-3 require less than 2 minutes. Additional cycles of the steps described above increasingly amplify the target DNA sequence. After the initial 5 minute cycle and a copy of the template DNA is made, the strands are reheated (reseparated) and then cooled, so another copying cycle can begin. Replicate cycles are run until the desired quantity of DNA of interest has been copied.

Figure 1.3 The polymerase chain reaction requires 1) Three main steps: Separation of dsDNA into ssDNA at 950 C, Primers binding to DNA template stands at 550 C, and formation of a new DNA strand (complementary to the template) using deoxynucleotides. 2-5) The cycle is repeated several times, yielding millions of copies of the target sequence in just hours.
One variation on this PCR process is "Hot Start." Hot start PCR starts the reaction with all of the ingredients except for taq polymerase. The mixture is run for 4 min. at 950 C, so when the taq is added, the solution is already hot and the primers have not had the chance to bind to one another. Now they are free to bind to the DNA sequence and help taq extend the new complemetary sequence.
Six main ingredients are necessary for a successful PCR procedure:
1) Water
2) Buffer ( 100 mM Tris-HCl, pH 8.3, 500 mM KCl, 1.5 mM MgCl2)4: maintains a pH in the DNA solution that is conducive to PCR.
3) Deoxynucleotides: components necessary to DNA synthesis (equal amounts of Adenine, Cytosine, Guanine, and Thymine deoxynucleotides should be used).
4) Primers: short DNA sequences that bind to ssDNA templates and allow polymerase to add deoxynucleotides.
5) Taq Polymerase: Heat resistant enzyme that adds deoxynucleotides complementarily to the DNA template strand. (Taq, isolated from Thermus aquaticus, allows the bacterium to survive at such high temperatures.)
6) Template DNA: DNA of interest which will be copied during the course of PCR.
References2:
1 Campbell, Neil. "Polymerase Chain Reaction." Biology. 4th ed. New York: Benjamin/Cummings Publishing Co.; 1996. p. 379.
2 Harnack and Kleppinger. Online!: a reference guide to using internet sources. New York: St. Martin's Press; 1997. p. 107.
3 National Center for Human Genome Research, National Institutes of Health. 1992. "New Tools for Tomorrow's Health Research: Polymerase Chain Reaction - Xeroxing DNA." www.gene.com/ae/AB/IE/PCR_Xeroxing_DNA.html. 15 February 1998.
4 Veilleux, Connie. "PCR Technology." www.com/ae/LC/SS/PS/PCR/PCR_technology.html. 15 February 1998.