Making A Lot Out of A Little: the Polymerase Chain Reaction
"PCR (polymerase chain reaction) is the most important new scientific technology to come along in the last hundred years."
-Mark R. Hughes, Deputy Director of the National Center for Human Genome Research, National Institutes of Health (Powledge 1998)
Hot Springs, Motorcycle Rides, and the Nobel Prize: a Background Look at the Polymerase Chain Reaction (PCR)
At first glance, it seems improbable that a stretch of California highway, a hot spring in the Yellowstone National Park, and the Nobel Prize in Chemistry have anything in common. Nevertheless, hidden behind a facade of lose connections is "an idea...so ingenious that it revolutionizes the way people ask questions" (Rhee 1998). The breakthrough development of PCR technology by scientist Kary Mullis was conceived in 1983 (Stryer 1995) as Mullis cruised the Pacific Coast Highway from San Francisco to Mendocino on his motorcycle. Ten years later, his idea won him the Nobel Prize in Chemistry (Rhee 1998).
The beauty of Mullis' idea lies in its simplicity. In essence, PCR mimics the mechanism most organisms use to copy their DNA ("Xeroxing DNA" 1992). The main obstacle of the PCR technique is its use of a wide variety of temperatures to perform DNA replication. Most naturally-occurring, temperature-dependent enzymes are not able to cope with this variety of temperatures. It is here that the organism, Thermus aquaticus and its associated, heat-stable DNA polymerase (Taq DNA polymerase) become important. This thermophilic bacterium found in hot springs in the Yellowstone National Park produces a DNA polymerase that can withstand the high temperatures used during the PCR procedure ("From Simple Ideas" 1998).
In essence, the PCR technique allows the rapid production of multiple copies of a target DNA sequence. In this sense, PCR amplifies a segment of DNA. The cyclical PCR procedure synthesizes copies of DNA very quickly as the products of one PCR cycle act as the templates for the next cycle ("From Simple Ideas" 1998). This technique is so efficient that a target sequence of DNA can be amplified a billion-fold over the course of several hours (McClean 1997).
Not only is this technique fast and efficient, it is also applicable to the work of scientists in a variety of disciplines. PCR helps researchers in fields ranging from human health (especially with respect to HIV detection), to forensic studies, to evolutionary relationships, to ecological studies and animal behavior (Powledge 1998). PCR's multitude of applications comes from its ability to amplify small fragments of DNA which are "lost" amongst the large background of the total nucleic acid ("From Simple Ideas" 1998). In fact, there is enough DNA in one-tenth of one-millionth of a liter (0.1uL) of human saliva (which contains a small number of shed epithelial cells) to use the PCR technique to distinguish a particular genetic sequence as human. Furthermore, there is enough DNA trapped in an 80 million year-old fossilized pine resin to amplify by PCR (Brown 1995). In short, very little DNA is needed to make PCR work.
"The Whole is Greater Than the Sum of the Individual Parts": the Pieces of This Ingenious Puzzle
The concept behind PCR is so simple because there are very few ingredients needed to amplify a segment of DNA:
The DNA to be amplified may come from a variety of sources ranging from human pathogens (HIV or herpes), to blood or semen from a crime scene, to the DNA of a 40 million year-old termite (Powledge 1998). Target sequences can be very small (discussed above) or very large (up to 10,000 base pairs) (Stryer 1995). The key element of DNA that makes the use of PCR applicable in almost all cases is that the genetic material of each living organism (plants, animals, bacteria, and viruses) consists of the same deoxynucleotide building blocks assembled in different combinations (Powledge 1998). DNA is a polymer of the four different nucleotides, each bound to a different base: adenine (A), guanine (G), cytosine (C), and thymine (T). What is of particular importance with respect to DNA is that A is complementary to T and G is complementary to C. Therefore, A will always pair with T and G will always pair with C. This complementation is the cornerstone of the PCR technique.
DNA polymerases (discussed below), whether human, bacterial, or viral cannot copy a chain of DNA without a short sequence of nucleotides to begin the process ("Xeroxing DNA" 1992). In essence, DNA polymerases need a free 3' -OH group to which another nucleotide can be attached (McClean 1997). Therefore, an oligonucleotide primer consisting of around 20 to 30 nucleotides is necessary (Brown 1995, Stryer 1995). In nature, special enzymes called primases are responsible for the synthesis of primers. The marvels of modern technology have supplanted these enzymes with the DNA synthesizer which produces oligonucleotide primers.
Of course, the oligonucleotide primer cannot be any combination of bases that make up DNA. More precisely, primers must be complementary to the nucleotide sequences on either side of the target sequence (Powledge 1998). This means that although the specific sequence of the target DNA does not need to be known, the sequence of the nucleotides on the 3' end of the DNA target sequence must be known in order to make PCR possible. These regions of the DNA template are called flankers and without them, PCR would never get started. Because DNA has two strands that run anti-parallel, two flankers and therefore two oligonucleotide primers are necessary.
As mentioned previously, DNA is a polymer of the four different deoxynucleotides (A, G, C, and T). In addition, these nucleotides have complementary partners (A and T, G and C). Therefore, to synthesize new DNA, these nucleotides have to be present. With the help of DNA polymerase, the correct complement bases can be matched.
Another important element of the dNTPs used in PCR is that they contain a triphosphate group. This portion of the dNTPs is crucial with respect to extending the new DNA chain. The energy released when these triphosphate bonds are broken is what powers the DNA synthesis mechanism.
DNA polymerase is an enzyme that adds deoxynucleotides to the 3' -OH end of a growing DNA strand (McClean 1997). Without this enzyme, no additional nucleotides can be added to the oligonucleotide primer.
The PCR technique requires that the reaction mixture undergo many changes in temperature, ranging from around 50 degrees Celsius to around 95 degrees Celsius (see below for more details). For most enzymes, this range of temperatures, in particular the high temperatures would mean inevitable denaturation. In fact, Mullis and other researchers experienced this problem during the development of the PCR technique. To combat this problem, the DNA polymerase had to be replenished after every repetition of the PCR cycle ("From Simple Ideas" 1998). This technical short fall has since been alleviated through the discovery of a bacterium, Thermus aquaticus, which resides in the hot springs of the Yellowstone National Park. This bacterium produces its own DNA polymerase (Taq DNA polymerase) which retains its activity even at high temperatures. Therefore, it only needs to be added to the PCR sample once, in place of adding it at the conclusion of every PCR cycle ("From Simple Ideas" 1998).
More recently, other sources of thermostable DNA polymerases have been discovered. Organisms that reside at the junction of two tectonic plates on the ocean floor also produce DNA polymerases that can withstand the high temperatures present during the PCR reaction. It has been found that the DNA polymerases from these organisms are even more heat-stable than the DNA polymerase from Thermus aquaticus and may eventually replace the Taq DNA polymerase in the PCR reaction (McClean 1997).
The PCR reaction must be carried out in a reaction buffer containing magnesium ions. The use of a buffer with a low concentration of magnesium ions results in mispriming and amplification of non-target sequences. Conversely, too high of a concentration reduces primer annealing and results in inefficient DNA amplification. Finally, thermostable DNA polymerases (like Taq DNA polymerase) are magnesium-dependent. Therefore, a precise concentration of magnesium ions is necessary to maximize the efficiency of the polymerase and the specificity of the reaction (Podzorski et al. 1997).
In the early days of PCR, three water baths at three different temperatures were used to change the temperature of the PCR mixture during the PCR cycle. The maintenance of three separate water baths proved inconvenient for two reasons. First, a technician had to monitor the temperature of the water in each bath. Additionally, the technician was responsible for moving the PCR mixture between the baths during the PCR cycle. Recent technological developments have done away with the need for constant attention during the PCR cycle. "Mr. Cycle" was the first automated PCR machine. It consisted of a multi-channel automated liquid handler and two aluminum blocks ("From Simple Ideas" 1998). This device monitors and regulates the temperature of the PCR mixture without the continual supervision of a technician.
Round and Round: the PCR Cycle
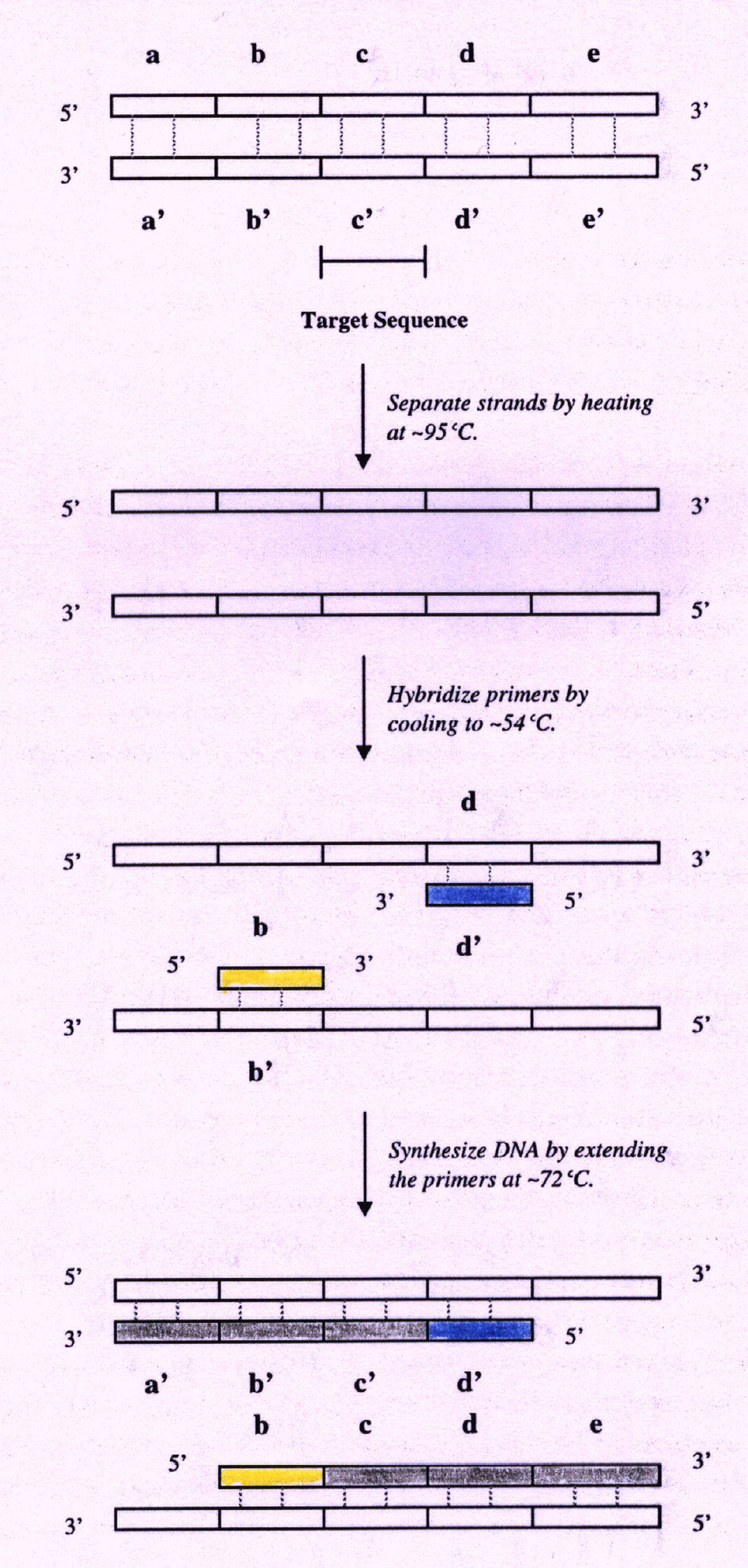
The PCR technique works through the repetition of a three-part cycle (Figure 1) (Brown 1995, McClean 1997, Powledge 1998, Stryer 1995). Each cycle consists of the following parts:
The two strands of the parent DNA (and of the target sequence) are separated by heating the PCR mixture to 95 degrees Celsius for 15 seconds (Stryer 1995). This is represented in Figure 1 by the breaking of the hydrogen bonds (dashed lines) between the DNA strands.
"Mr. Cycle" abruptly cools the PCR mixture to around 54 degrees Celsius. The temperature of this step is highly variable, depending on the size of the primer and its homology to the target DNA (McClean 1997). The primers are permitted to hybridize between 15 seconds and 2 minutes (Podzorski et al. 1997). In Figure 1, the two primers, d' and b attach to the flanker regions surrounding the target sequence. The primers are added in excess to prevent the formation of DNA duplexes (Stryer 1995).
Elongation of both primers now occurs through the activity of the Taq DNA polymerase. The reaction mixture is heated to 72 degrees Celsius, the optimal temperature for Taq DNA polymerase activity. Elongation of both primers occurs in the direction of the target sequence because the 3' ends of both primers face the target sequence. This portion of the cycle proceeds for between 15 seconds and 3 minutes (Podzorski et al. 1997).

Figure 1. Each cycle of PCR consists of three steps (Denaturation, Annealing, and Primer Extension) shown here schematically. Each part of the cycle is driven by changes in temperature. This cycle is repeated between 25 and 45 times (image based on Stryer 1995).
At the conclusion of the first cycle, both strands of the target have been replicated and can now act as templates during subsequent PCR cycles. In addition, more than just the target sequence has been replicated (Stryer 1995). Subsequent repetitions of the cycle not only increase the amount of DNA, but also refine the amount of target DNA that is being replicated. In Figure 1, for example, b-c-d-e from the first cycle serves as a template for the synthesis of b'-c'-d' during the second cycle. Similarly, a'-b'-c'-d' from the first cycle serves as the template for the synthesis of b-c-d during the second cycle. In the end, DNA consisting of the target sequence and the flanking sequences increases exponentially (2 to the n power) while the other DNA sequences (which contain more than the target sequences and flankers) increase linearly (Stryer 1995). Figure 2 shows how repeating cycles create shorter pieces of DNA more readily than the larger fragments.
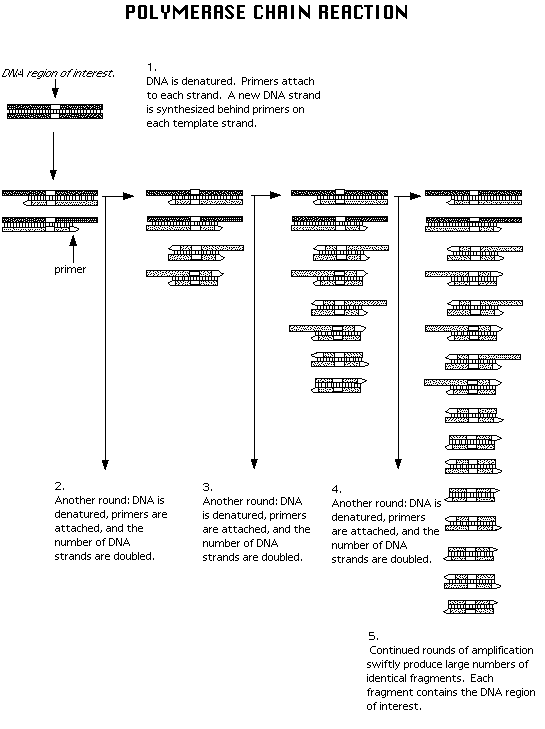
Figure 2. Multiple cycles of PCR produce many copies of the DNA target sequence and flankers while longer sequences (target sequence, flankers, and additional DNA) increase at a slow rate ("Polymerase Chain Reaction." Biotech Graphics Gallery. 1998. <http://www.gene.com/ae/AB/GG/polymerase.html> (01 February 1998)).
A variable number of repetitions (25-45) can be executed (Podzorski et al. 1997). In the end, the specific target sequence can be amplified between one million and one-billion fold (Powledge 1998).
For more information about PCR, go to the PCR Jump Station: "the ultimateWeb page for information and links on all aspects of the Polymerase Chain Reaction (PCR)." Here you can link to just about any information about PCR that might interest you from catalogs to protocols to general information.
Works Cited
Biotech. 1998. "Polymerase Chain Reaction (PCR)." Biotech Graphic Gallery <http://www.gene.com/ae/AB/GG/polymerase.html> Accessed 01 February 1998.
Biotech. 1998. "Polymerase Chain Reaction - From Simple Ideas." Biotech Issues & Ethics. <http://www.gene.com/ae/AB/IE/PCR_From_Simple_Ideas.html> Accessed 01 February 1998.
Biotech. 1992. "Polymerase Chain Reaction - Xeroxing DNA." Biotech Issues & Ethics. <http://www.gene.com/ae/AB/IE/PCR_Xeroxing_DNA.html> Accessed 01 February 1998.
Brown, J.C. 1995. "What the Heck Is PCR?" <http://falcon.cc.ukans.edu/~jbrown/pcr.html> Accessed 01 February 1998.
McClean, P. 1997. "Polymerase Chain Reaction (or PCR)." Cloning and Molecular Analysis of Genes. <http://www.ndsu.nodak.edu/instruct/mcclean/plsc431/cloning/clone9.htm> Accessed 01 February 1998.
Podzorski, R.P., Kukuruga, D.L., Long, M.P. 1997. "Introduction to Molecular Methodology." In: Manual of Clinical Laboratory Immunology, 5th edn. Noel R. Rose, Everly Conway de Macario, James D. Fold, H. Clifford Lane, Robert M. Nakamura, editors. Washington, D.C.: American Society for Microbiology, 77-107.
Powledge, T.M. 1998. "The Polymerase Chain Reaction." Breakthroughs in Bioscience. <http://www.faseb.org/opar/bloodsupply/pcr.html> Accessed 01 February 1998.
Rhee, S.Y. 1998. "Kary B. Mullis (1945- )." Biotech. <http://www.gene.com/ae/AB/BC/Kary_B_Mullis.html> Accessed 01 February 1998.
Stryer, L. 1995. Biochemistry, 4th edn. New York: W.H. Freeman and Company.