MacDNAsis of Hexokinase
MacDNAsis (version 3.5) is a computer program that allows a cDNA sequence to be analyzed. It will determine where the code for the protein is located on the cDNA, the molecular weight of this protein and other physical characteristics of the protein from one cDNA sequence. The program then will compare the amino acid sequence for this protein from different species and determine the similarity between the species. The first five steps of analysis of hexokinase use only cDNA which was obtained from S.cerevisiae (yeast). To see the original information click on the below picture.
The first step of MacDNAsis was to determine what part of the cDNA encoded for the protein hexokinase (figure one). This portion of DNA is called the Open Reading Frame. It begins with the start codon ATG, which also codes for methionine, and ends with the stop codon. The picture below illustrates the cDNA of yeast. The red triangles indicate the start codons, the green lines indicate stop codons. The black section was chosen as the open reading frame since it is the largest section between a start and stop codon. The largest ORF is between nucleotides 718-2178 which encodes for 487 amino acids that results in a 53,949.57 Dalton protein. From examining this picture, there seems to be a large section of 3' untranslated for hexokinase.
Figure 1: Open Reading Frame of Hexokinase
The next step in analysis was to translate the open reading frame. Once the computer program had done this, the resulting amino acid sequence was analyzed. Below is a picture of the Kyte & Doolittle hydropathy plot (figure 2). A hydropathy plot measure the hydrophobicity for a certain section of protein. The section of the protein is indicated by the window number. In this plot, a window of 12 amino acids was used meaning that the computer looked at a sequence of twelve amino acids and calculated the hydrophobicity of those twelve. The greater the positive number, the more hydrophobic that portion is. A hydropathy plot is used to determine if there are potential transmembrane domains in the protein. A potential transmembrane domain is > 1.8 on the scale. It seems as if hexokinase has one potential transmembrane domain located at amino acid 390. Therefore, hexokinase is most likely not an integral membrane protein in yeast. However, there is a possiblitiy that hexokinase would span the membrane once.
Figure 2: Hydropathy Plot
Hexokinase was also analyzed for its antigenicity. This was done with a Hopp & Woods plot that also had a window of 12 amino acids (figure 3). Antigenicity means the area of the protein that would most likely be able to generate a monoclonal antibody from that peptide sequence which would recognize the protein in its native conformation. This is useful to know in order to make a probe that binds to the protein. The plot measures antigenicity by examing the hydrophylcity of the protein. A hydrophilic section of the protein would most likely be located in the cytoplasm of the cell and be an available epitope. From the plot below, it seems like the regions around 250, 300, 350 and 380 amino acids would be good epitopes for a monoclonal antibody because of the high values on the plot. However, the Hopp & Woods plot is only a predictor and can be wrong.
Figure 3: Antigenicity Plot
The computer generated amino acid sequence of hexokinase which is the promary structure of the protein. From this, a model for the secondary structure was created (figure 4).
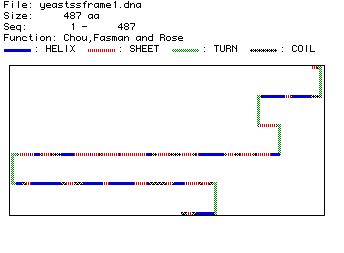
Figure 4: Secondary Structure of Hexokinase in Yeast
This prediction shows the majority of hexokinase to be made up of beta pleated sheets with an occasional alpha helix breaking up the sheet. Also, hexokinase is predicted to have five turns. When compared with the rasmol image of hexokinase when divided into different structures, it seems consistent because the alpha helixes seem to be before and after the beta pleated sheets, with a small helix interupting three or four times as is predicted. Below is a picture of hexokinase divided into structures and in ribbons. (Pink = alpha helix, yellow = beta pleated sheets)
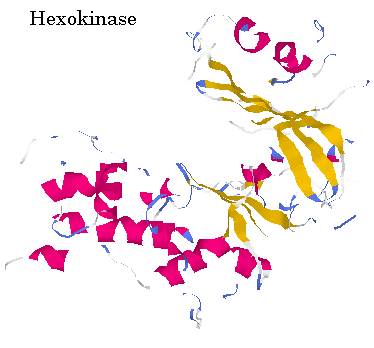
Figure 5: RasMol snapshot of Hexokinase
The amino acid sequence of hexokinase was compared for five different organisms: yeast, blood fluke, mouse, cow and human. To go see the Genbank information on one of these organisms, click on their name below.
Below is picture that shows the homologous amino acids (single letter abbreviation) for a portion of the sequence (figure 6). This was generated through the Higgins method. Dashed lines are used to maximize the alignment of the amino acids. This is necessary because hexokinase is a different length in different organisms. Black boxes around the letters show conserved amino acids.
Figure 6: Homology of Hexokinase Protein
A phylogenic tree is much easier to read and determine the similarity of hexokinase in different organisms (figure 7). Human and mice hexokinase is the most similar which would indicate that the proteins diverged the most recently. It seems that cow hexokinase split a little earlier in its evolution because of the lower percentage of similarity. Blood fluke and yeast hexokinase must have diverged a long time ago in order to account for the very lower percentage similarity.
Figure 7: Phylogenic Tree of Hexokinase
Return
to My Home Page