This page was created for an undergraduate Molecular Biology course at Davidson College.
1) Glycine max - Eukaryote
2) Homo sapiens - Eukaryote
3) Mouse - Eukaryote
4) Rat - Eukaryote
5) Arabidopsis thaliana - Eukaryote
6) E. coli - Prokaryote
In studying the structure of IDH, a particularly important paper is Panisko et al., 2001. We learn that IDH operates as an octamer of four IDH1's and four IDH2's. IDH1 performs regulatory funcitons and IDH2 contains the catalytic site. IDH1 is regulated by both isocitrate and AMP. This paper is interested in learning how and where the two subunits interact with one another in forming a functional octamer complex. In order to explore this, it is helpful that we have available the ortholog of IDH in E. coli, because Panisko et al use the three dimensional structure of E. coli IDH proteins (Chime page contains almost exclusively E. coli IDH images) to learn about IDH1-IDH2 interactions in yeast. In the homologous E. coli protein it appears that the IDH1 and IDH2 proteins have binding interaction at the isocitrate binding site. It was previously known that amino acids Lys-183 and Asp-217 on IDH1 played a role in the catalytic site of IDH2. Panisko et al ran an experiment in which they replaced these amino acids with alanine. The result is a lowered catalytic function of the enzyme. This implies that these two amino acids in IDH1(Lys-183 and Asp-217) in yeast do in fact play a role in the catalytic site of IDH2 which when mutated reduces the overall catalytic function of the complex. Panisko et al then tested to see if changing two amino acids (Lys-189 and Asp-222) on IDH2 will have an affect on the regulatory binding site of IDH1. Mutating either of these amino acids prevented the complex from being activated by AMP. Also, mutating these two amino acids affected the cooperative binding of isocitrate.
These experiments were very well conceived. The team took previous knowledge of IDH1 and IDH2 working as a heterodimer and combined it with information on the 3D structure of homologous proteins in E. coli. They then tested to see if changes that affect the model organism will also affect yeast. Given the results to both mutation experiments (on IDH1 then IDH2) they show that each subunit of the heterodimer of IDH1/IDH2 affects the isocitrate binding site of the other.
Another particularly interesting structure paper is Hurley et al, 1996. This paper also makes use of the 3D structure of E. coli IDH proteins (note in particular Chime 5). What this paper performs is a switch in binding affinity of an IDP mutant from coenzyme NADP+ to NAD+ (if it started binding to NADP+ then I assume it was an IDP, though not specifically stated in the paper). This is performed by making a number of amino acid changes which alter the structure of the moleculre. As is always the case, funtion follows form. In this case, they made just the right adjustments in order that the protein switched its coenzyme of choice. Check out the chime images under Chime 5 in sequence paying careful attention to the how the changes made by the scientists affect the binding of the molecule.
Yet another in the seeemingly endless sea of cool IDH structure papers is Doyle et al, 2001. This paper begins by stating that a WT IDH protein complex is quite specific in binding isocitrate, and not other "(R)-malate-type substrates". They have found a mutant with a binding specificity switched from isocitrate to isopropylmalate. In order to determine structural changes in this mutant allow it to bind a different substrate, they created 3D image of the IDH mutant (see Chime 3). They learned that there were changes in the global structure and near the active site. In particular they refer to the loop and helix near the active site being changed. This switch affects the binding of the substrate NADP+. These changes in concert lead to the change in specificity of the IDH from isocitrate to isopropylmalate.
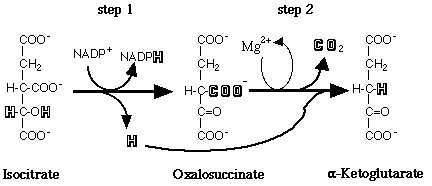
"Isocitrate dehydrogenase (IDH) catalyzes an oxidation-reduction reaction in which isocitrate is converted to a-ketoglutarate (2-oxoglutarate) and CO2 (Figure 1)"

Figure 1: Conversion of isocitrate to a-ketoglutarate via degygogenation and decarboxylation by IDH.
*This figure was taken from the introduction of Lowell Y.M. Rayburn's Honor Thesis available online.
Panisko EA, McAlister-Henn L. 2001. Subunit interactions of yeast NAD+-specific isocitrate dehydrogenase. J Biol Chem. 276(2):1204-10
Hurley JH, Chen R, Dean AM. Determinants of cofactor specificity in isocitrate dehydrogenase: structure of an engineered NADP+ --> NAD+ specificity-reversal mutant. Biochemistry 1996 May 7;35(18):5670-8
Doyle SA, Beernink PT, Koshland DE Jr. Structural basis for a change in substrate specificity: crystal structure of S113E isocitrate dehydrogenase in a complex with isopropylmalate, Mg2+, and NADP. Biochemistry 2001 Apr 10;40(14):4234-41
Rayburn, Lowell. 1999. Isocitrate Dehydrogenase in Drosophila melanogaster: One Subunit or Two? Honors Thesis, Davidson College
Contact Kevin James with any comments or questions