This page was created as part of an undergraduate course at Davidson College.
Checking for Orientation of IDH Gene Insert in a Plasmid Vector
After cloning your favorite gene into a plasmid, it is still unknown whether the gene ligated in correctly or whether the insert ligated in upside down and backwards. One can check for the correct orientation of their favorite gene in a plasmid by restriction digesting. Using a restriction enzyme to cut at a known location in your favorite gene insert and somewhere else on the plasmid allows one to predict the sizes of the cut fragments, which will differ if your favorite gene was inserted correctly or incorrectly. Here's an example using the IDH1 gene inserted in the pQE 30 UA plasmid vector.
IDH1
1. Select restriction enzymes to digest plasmid using a restriction map for IDH1 and a restriction map of the plasmid vector you are using. The restriction enzyme should cut a a known spot in the IDH1 insert and a known place somewhere else in the vector.
2. From these data, you can predict the size of the cut fragments produced from the restriction digestion. Using the restriction enzyme ECO RV, the plasmid is cut at basepairs 340 and 743; this is 16 bases upstream of the gene insert. So, if the gene is inserted correctly it would be a fragment 340+16=356bp in length or if it was inserted incorrectly it would be a fragment 743+16=759bp in length.
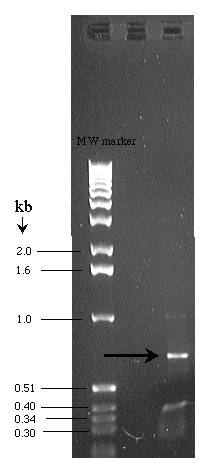
3. Once you know the expected size of the correctly and incorrectly inserted gene, you can run your restriction enzyme on a gel to determine the actual size of the cut fragments in the restriction digestion, which will appear as bands of different sizes. Compare the bands of your restriction digestion in the gel to the known sizes of a molecular weight marker, and you know both the size of the fragments in your restriction digestion and whether your favorite gene was inserted correctly.
This is an example of a sequence fragment run on a gel next to a molecular weight marker for comparison.
IDH2
Restriction Digestion:
Using ECO RV to digest IDH2.
Correct Insertion fragment: 790+16=806 bp
Incorrect Insertion fragment: 320+16=336 bp
IDP1
Restriction Digestion:
Using Bam HI to digest IDP1.
Correct Insertion fragment: 780+29=809 bp
Incorrect Insertion fragment: 507+29=536 bp
IDP2
Restriction Digestion:
Using BGL II to digest IDP2.
Correct Insertion fragment: 743+15=758 bp
Incorrect Insertion fragment: 496+15=511bp
IDP3
Restriction Digestion:
Using Cla I and Bam HI to digest IDP3 (Cla I cuts the plasmid and Bam HI cuts the gene insert).
Correct Insertion fragment: 495+29=524 bp
Incorrect Insertion fragment: 768+29=797 bp
Contact: Peter Leese or view Molecular Biology at Davidson