This web page
was created as assignment for an undergraduate course at Davidson College.
structure & orthologs
|
Isocitrate dehydrogenase, or IDH, has five isoforms: IDH1, IDH2, IDP1, IDP2, and IDP3. These isoforms were discussed in the last assignment in the context of cloning the genes into a plasmid. In order to learn basic information regarding the nucleotide sequence, amino acid sequence, and restriction sites of these five isoforms, please view assignment 1.
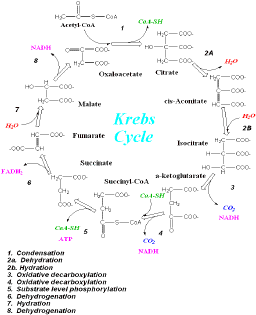
This enzyme is involved in Step 3 of the Citric Acid Cycle (Krebs Cycle), the first of the two oxidative decarboxylation steps (Figure 1).
|
|
|
Figure1.
Krebs Cycle. Step 3 involves the conversion of isocitrate to a-ketoglutarate.
Original figure taken from http://wine1.sb.fsu.edu/krebs/krebs.htm. |
Isocitrate dehydrogenase catalyzes the oxidation of D-Isocitrate to a-Ketoglutarate and carbon dioxide (Figure 2). A cofactor such as nicotinamide adenine dinucleotide (NADH) or nicotinamide adenine dinucleotide phosphate (NADPH) is required for this decarboxylation. NAD+ and NADP+ are reduced in the mitochondria and cytosol, respectively (http://wine1.sb.fsu.edu/krebs/krebs.htm).
|
|
|
Figure 2.
First Oxidative
Decarboxylation Step of the Krebs Cycle (Citric Acid Cycle). The oxidation of isocitrate to a-ketoglutarate
occurs in the presence of IDH and a cofactor (NAD+ or NADP+). Original figure taken from http://wine1.sb.fsu.edu/krebs/krebs.htm. |
Information regarding protein structure and orthologs can be applied to my molecular biology class project. My class first set out to clone all five IDH isoforms into a plasmid vector. We are currently running Southern Blots to confirm the presence of the inserts and to ensure that the inserts are the intended isoform. Next, my class would like to express the isocitrate dehydrogenase proteins in bacteria. Information regarding protein structure and orthologs may prove to be helpful in accomplishing this feat and thus should be exploredÍ
|
structure |
Isocitrate dehydrogenase structure is shown in the following Chime figures. Structural differences are evident when viewing IDH complexing with nicotinamide adenine dinucleotide (NADH) or nicotinamide adenine dinulceotide phosphate (NADPH).
Isopropylmalate dehydrogenase (PDB)
I could not find a chime image of isocitrate dehydrogenase complexed with NAD+, so I opted for an enzyme that is evolutionarily related (Karlstrom et al. 2000) and orthologous (Table 1) to isocitrate dehydrogenase (Hurley, 1994).
Isocitrate
dehydrogenase complexed with Isocitrate, NADP+, and Calcium (Flash-cooled)
(PDB)
dimer from Esherichia coli
(Mesecar et al. 1997)
|
orthologs |
Proteins orthologous to IDP2 in Saccharomyces cerevisiae were determined using a protein-protein BLAST on NCBI.
Table
1. Orthologs
of IDP2. Eukaryotic and prokaryotic
proteins that are orthologous to IDP2 in Saccharomyces cerevisiae.
|
kingdom |
organism |
protein |
E value |
|
Eukaryote |
Homo sapiens
|
isocitrate dehydrogenase |
e-132 |
|
Eukaryote |
isocitrate dehydrogense
2 |
e-145 |
|
|
Eukaryote |
GH12815P |
2e-5 |
|
|
Eukaryote |
isocitrate &
isopropylmalate dehydrogenases |
e-144 |
|
|
Prokaryote |
3-isopropylmalate
dehydrogenase |
3e-6 |
|
|
Prokaryote |
icd1 |
e-141 |
By analyzing the E values, one can infer that both prokaryotes and eukaryotes share significant sequence similarity with the IDP2 protein of Saccharomyces cerevisiae. (The smaller the E value, the less chance there is for random sequence similarity.) In fact, IDH is so highly conserved that it is found in species that may not even have a citric acid cycle (Karlstrom et al. 2000). Karlstrom et al. also divulge that isopropylmalate dehydrogenase is evolutionarily similar to isocitrate dehydrogenase, a fact apparent in Table 1, where both C. elegans and C. crescentus orthologs are significantly structurally similar to the yeast IDP2 gene. The house mouse (Mus musculus) and C. elegans demonstrate the greatest sequence similarity with E values of 10-145 and 10-144, respectively.
|
laboratory
significance |
In lab, Liz Shafer and I have worked to clone the IDP2 gene of Saccharomyces cerevisiae into a plasmid vector, pQE-30UA. We attempted to create eight inserts, however only one bacterial colony inserted the yeast DNA. We used restriction endonuclease digestions and gel electrophresis to determine the orientation of the insert. The results indicated a reverse orientation, which would not allow for the expression of the appropriate protein since the mRNA would be transcribed in reverse. Only a short protein would be transcribed that was comprised of the 6-His tag and amino cids up until a stop codon. The chime figures illustrate what the IDH protein may have looked like if we were able to correctly express it.
|
references |
Karlstrom, M et al. (2000) Isocitrate dehydrogenase from Archaea.
http://www.csb.ki.se/xray/idh.html