This web page was produced by Zack Perfect as an assignment for an undergraduate course at Davidson College
Determining Orientation of Plasmid
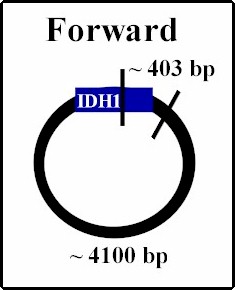
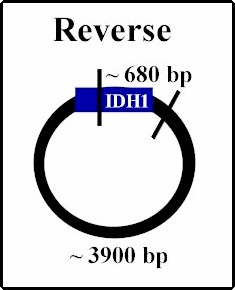
| Once PCRed, the gene of interest IDH1, is ligated into the plasmid. Because the sticky edges of the plasmid are the same, being cut with the same restriction enzyme, the gene could insert into the plasmid either right side up (forward) which is desired, or upside down (reverse). In order to determine the orientation we took samples from various colonies of E. coli we grew containing different possible insert orientations. We used restriction enzyme that cut once inside the gene and once outside, therefore producing differing sizes of DNA strands depending on the orientation. We used Sal I; below are predictions of the framents under the two different scenarios. When the digestions are run on gel electrophoresis, two bands should appear and by determining their sizes, the orientation should be confirmed. The samples containing forward orientation can be used to proceed with the experiment. |
Digested with Sal I


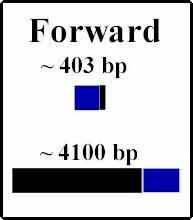

Other predicted fragment sizes
IDH2 cut with EcoRV Forward: ~ 320 bp Reverse: ~ 790 bp
IDP1 cut with BamHI Forward: ~ 507 bp Reverse: ~ 780 bp
IDP2 cut with BglII Forward: ~ 743 bp Reverse: ~ 496 bp
IDP3 cut with Cla I then BamHI Forward: ~ 522 bp Reverse: ~ 795 bp