This web page was created as an assignment for an undergraduate course at Davidson College.
Structure and Orthologs:
Isocitrate Dehydrogenase
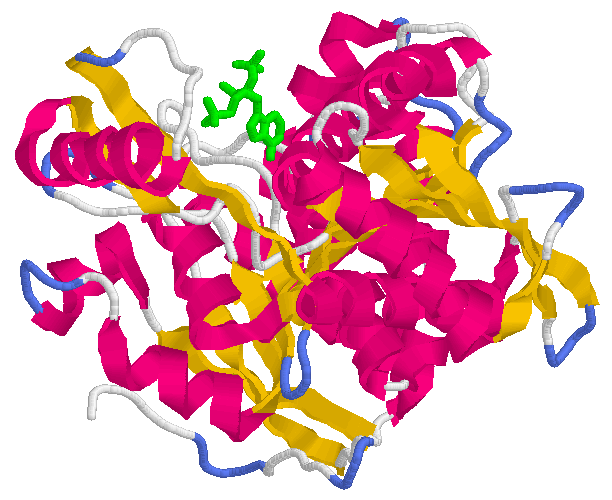
E. coli isocitrate dehydrogenase with NADP(+) (green). Click on picture to go to source. Authors: J. H. Hurley, A. M. Dean, D. E. Koshland Jr, R. M. Stroud Reference: Biochemistry, 1991, 30, 8671.
Isocitrate dehydrogenase, IDH, is a protein that catalyzes a reaction in the citric acid cycle, giving the cell both NADPH and 2-oxoglutarate, two molecules needed for the synthesis of specific lipids and amino acids (Purves et al., 1998). The NAD(+)-dependent IDH in Saccharomyces cerevisiae is composed of two subunits, IDH1 and IDH2, that are coded by two separate genes (also designated IDH1 and IDH2). IDH1 contributes regulatory properties to the complex and IDH2 contains the catalytic site (Panisko et al., 2001). The NADP(+)-specific isocitrate dehydrogenases are the cytosolic isozyme IDP1, the mitochodrial isozyme IDP2, and the peroxisomal isozyme, IDP3 (Loftus et al., 1994 and Henke et al., 1998).
NAD-dependent isocitrate dehydrogenase and NADP-dependent isocitrate dehydrogenase are present in both prokaryotes and eukaryotes and involved in the generation of both NADP(+) and alpha-ketoglutarate for biosynthetic pathways (Chen et al., 2000). The entire paper by Chen et al. is extremely beneficial in looking at orthologs of IDH and the evolutionary impact on these genes.
To see isolated three-dimensional structures of IDH proteins from different species, see Chime Images
To see proteins similar in sequence to the yeast IDH2 protein, click Orthologs
© Copyright 2002 Department of Biology, Davidson College, Davidson NC 28035
Send comments, questions, and suggestions to vistatler@davidson.edu