This web page was produced as an assignment for an undergraduate course at Davidson College.
In Situ Hybridization
What is In Situ Hybridization?
In Situ Hybridization (ISH) uses a labeled probe to detect and localize
specific RNA or DNA sequences in a tissue or on a chromosome. ISH relies on
DNA's ability to reanneal, or hybridize, with a complimentary strand when at
the correct temperature. “In situ” means “in the
original place” in Latin, so ISH involves a labeled nucleic acid probe
hybridizing with a DNA or RNA sequence in situ (in the cells) so that
the location of the sequence of interest can be detected in the cell, tissue,
or chromosome. (Immuno-ISH, http://home.no.net/immuno/).
Like Northern and Southern Blots, ISH indicates the presence of a particular
RNA or DNA sequence, but ISH differs from blots in that the labeled probe reveals
the actual location of the sequence in the cells. ISH is the only procedure
that allows the location of the sequence of interest to be studied (Polak et
al., 1990). The probe can be either radioactively labeled and detected
by autoradiography or fluorescently labeled (abbreviated FISH) and detected
by immunocytochemistry. The specificity of the probe depends on the permeability
of the cells, the type of probe, the labeling technique, and the hybridization
conditions, so specificity of ISH can be adjusting according to the desired
results (Polak et al. , 1990).
How does ISH work?
The general procedure for ISH involves fixing samples of chromosomes or tissues
onto a glass slide. The material is then treated with chemicals to permeabilize
the cells or tissues and denature the DNA so that the probe can more readily
hybridize. A complementary nucleic acid probe is prepared and labeled, either
by radioactivity or by fluorescence. A hybridization solution containing the
probe is sloshed over the slide so that the probe can hybridize with the sequence
of interest. The excess probe is washed away and then either autoradiography
or immunocytochemistry is used to detect the location of the probe. Microscopy
can be used to view the samples (Leitch et al., 1994). See Figure
1 for a flow chart of FISH.

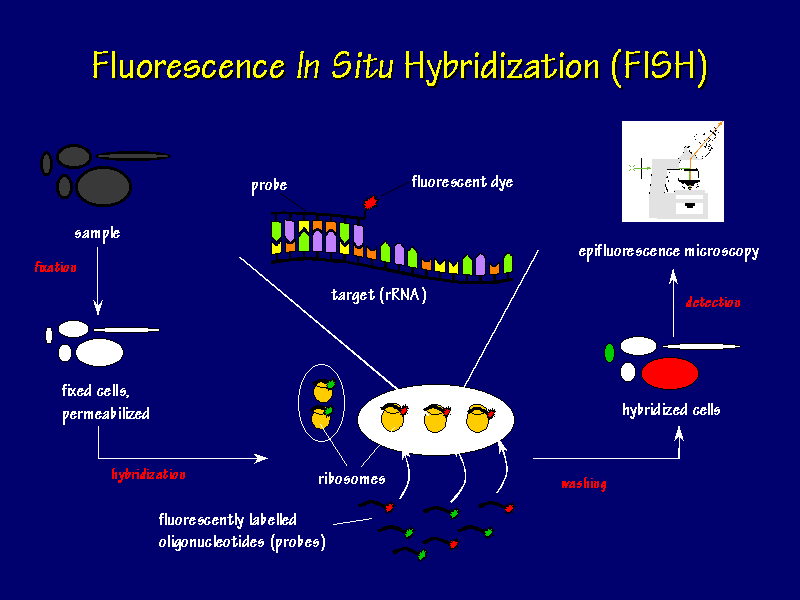
Figure 1. Fluorescent In Situ Hybridization Flow Chart. The samples are fixed onto coverslips, fluorescently labeled oligonucleotide probes are hybridized with the sample, excess probes are wahed away, and epifluorescence microscopy is used to view the location of the hybrized cells. (Permission from Dr. Frank Oliver Glockner for use of image found at http://www.mpi-bremen.de/molecol/fog/fish1.html.)
Fixation and Denaturation of Material
The DNA or RNA must be fixed onto a glass slide for stability. so as to allow
the probe sufficient access to the targe maintain the structure as much as possible
while giving the probe sufficient access to the target by permeabilizing the
cells. While DNA is relatively stable and can be fixed easily, RNA is highly
degradable, so extra care must be taken to fix the mRNA onto the slide as soon
as possible after it is extracted (Polak et al., 1990). Possible fixatives
include paraformaldehyde, formalin, or paraffin embedding. Cells must be treated
with detergent or proteinases such as triton and RNAse-free proteinase K in
order to permeabilize the membranes. The probe must have sufficient access into
the cells so that it can bind with the target. The degree to which the cells
are permeabilized affects the degree of specificity of ISH (Immuno-ISH, http://home.no.net/immuno/).
Probes
A variety of probes can be developed for ISH, depending upon the desired binding
sensitivity. For DNA ISH, probes include double-stranded DNA, single-stranded
DNA, and synthetic oligodeoxyribonucleotides. For RNA ISH, the probe can be
single-stranded complementary RNA, called a riboprobe, that is synthesized by
reverse cloning (Polak et al., 1990). Double-stranded DNA probes can
be produced by PCR or replicated in bacteria. Double-stranded DNA probes must
be denatured before hybridization and are often less sensitive because the single
strands of DNA have a tendency to reanneal with each other. Single stranded
DNA probes can be produced by PCR or reverse transcription of RNA. Oligonucleotide
probes are synthetically produced and are relatively small (20-40 basepairs).
Oligonucleotide probes can fit easilty through permeabilized cell membranes
and can be designed to be very specific. Complementary RNA probes can be produced
from RNA. RNA-RNA interactions are very stable and resistent to RNAses, however
before hybridization they are difficult to work with (Power and Versatility
of ISH, http://martin.parasitology.mcgill.ca/insituhybridization/INDEX.HTM).
Once the probe is developed, it must be labeled so that the location of the target sequence can be detected. Probes can be labeled directly, where the reporter molecule is directly attached to the probe, or indirectly, where a specific antibody or labeled binding protein is used to detect another molecule that is attached to the probe sequence. Radioactively labeled probes were originally used for ISH and continue to be used because they can be synthesized and incorporated into the DNA or RNA easily and autoradiography is relatively sensitive (Polak et al., 1990). However, radioactively labeled probes have a limited shelf life and require additional safety procedures and autoradiography can take several days. Non-radioactively labeled probes (fluorescently labeled probes) are very popular, as the procedures are readily available and less time consuming (Immuno-ISH, http://home.no.net/immuno/). Typical radioactively-labeled probes use 32P or 35S isotopes. Fluorescin and Rhodamine can be used for direct fluorescently labeled probes, and biotin and digoxigenin can be used for indirect fluorescence labeling. Fluorescent labeling can allow two or more different probes to be visualized at the same time because of color differences (Power and Versatility of ISH, http://martin.parasitology.mcgill.ca/insituhybridization/INDEX.HTM).
Hybridization
The degree of specificity to which the probe hybridizes to the target sequence
can be controlled by the design of the probe and the conditions of the buffer
solution, including temperature, pH, and salt concentration. “High stringency”
conditions will only allow hybridization of probes with very similar homology
to the target sequence, while “low stringency” conditions will allow
a probe to bind with less specificity. Hybridization mixtures usually have a
small volume (about 10-20 ul total) with 50% formamide and hybridization typically
occurs between 37-60 degrees Celsius (Polak et al., 1990).
Visualization
Several different techniques can be used to view where the probe has hybridized
with the sequence of interest. Light field microscopy is most common and can
be used for radioactively labeled probes or probes labeled with peroxidase or
alkaline phosphatase. Fluorescent microscopes are used to view fluorescently
labeled probes; the UV light excites the fluorescent die so that it can be detected
through the microscope. Digital imaging systems are also used and can process
the images and do quantitative measurements (Power and Versatility of ISH, http://martin.parasitology.mcgill.ca/insituhybridization/INDEX.HTM).
See Figure 2 for an example of a FISH image.
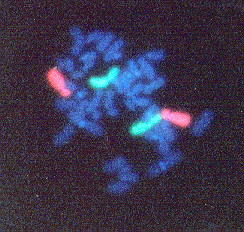
Figure 2. FISH performed on chromosomes from a human peripheral blood lymphocyte. Chromosome 2 pair is labeled green and the Chromosome 4 pair is labeled orange. This image shows an example of how FISH can be used to identify specific pairs of chromosomes. (Permission pending for image found at http://www1.na.infn.it/wnucl/lines/radi/fish2.html.)
Experimental Controls
Like any good experiment, controls must also be developed for ISH to make sure
that the probe is specific and binds only to the sequence of interest and that
all steps of the procedure have been carried out properly. Controls for RNA
In Situ Hybridization can include a poly-T tail to determine how strong
of a signal is being detected (Power and Versatility of ISH, http://martin.parasitology.mcgill.ca/insituhybridization/INDEX.HTM).
Northern and Southern blots can also be used to determine the strength of the
signal for RNA and DNA. For apositive control for RNA, a probe for a constituitively
expressed gene such as actin will demonstrate the procedure is working correctly.
For a positive control for DNA, a probe should be synthesized for a highly repeated
sequence (Leitch et al., 1994).
Applications of ISH
ISH is used in medical research and diagnosis, biology, pathology, and plant
breeding. ISH can be modified in a variety of ways depending upon the material
being analyzed. Some of ISH applications include determination of chromosome
structure, function, and evolution, chromosomal gene mapping, expression of
genes, localization of viral DNA sequences, diagnosis of viral diseases, and
localization of oncogenes, sex determination. The uses and different approaches
for ISH continue to increase, thus impacting many different research fields.
New labeling techniques for probes, new detection systems, and advanced computer
software increase the availability and efficiency of ISH (Leitch et al.,
1994).
Brief History
ISF was first introduced in 1969 independently by Gall and Pardue (1969), Buongiorno-Nardelli
and Amaldi (1969), and John et al. (1969). Radioactively labeled RNA
was used to detect DNA in vivo. Tritium was incorporated into proliferating
cells in order to label the newly synthesized nucleic acids. While the labeling
techniques and probe design have changed slightly, the basic technique of ISH
remains the same today (Polak et al., 1990).
Related Sites
http://martin.parasitology.mcgill.ca/insituhybridization/INDEX.HTM
This site provides a good and clear overall description of In Situ
Hybridization. It also has a cool slide show.
http://home.no.net/immuno/
This site provides detailed information about ISH, along with information about
immunofluorescence and ISH procedures.
http://www.ksu.edu/wgrc/Protocols/Cytogenetics/ISH.html
This site gives several sample protocols for ISH.
http://www.findarticles.com/cf_dls/m2250/9_38/55812259/p1/article.jhtml
This site talks about FISH involved with clinical testing.
References
Glockner, Frank Oliver, 2002. Fluorescence In Situ Hybridization. <http://www.mpi-bremen.de/molecol/fog/fish1.html
>. Accessed 2003 Feb. 18.
Immunohistochemistry-In Situ Hybridization. In Situ Hybridization. <http://home.no.net/immuno/>. Accessed 2003 Feb. 16.
Leitch, A. R. et al. 1994. In Situ Hybridization. BIOS Scientific Publishers Limited, Oxford, pp. 1-33.
Polak, Julia M. and James O’D McGee. 1990. In Situ Hybridization: Principles and Practice. Oxford University Press, Oxford, pp. 10-30.
The Power and Versatility of In Situ Hybridization. In Situ Hybridization Lecture. < http://martin.parasitology.mcgill.ca/insituhybridization/INDEX.HTM>. Accessed 2003 Feb. 16.
Radiation Biophysics Laboratory. Flourescence In Situ Hybridization. <http://www1.na.infn.it/wnucl/lines/radi/fish2.html >. Accessed Feb. 18, 2003.
__________________________________________________________________________________
Davidson College Biology Department
Davidson College Molecular Biology
Send questions and comments to Sarah Baxter at sabaxter@davidson.edu.
Thanks!