*This website was produced as an assignment for an undergraduate course at Davidson College.*
Creating a cDNA Library from mRNA Isolated Using the Oligo(dT) Column
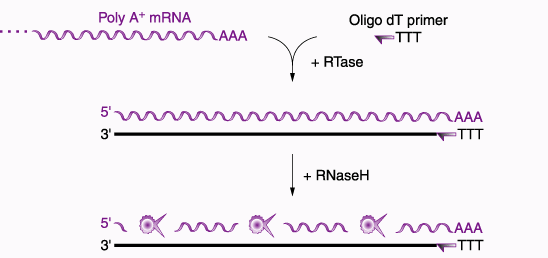
After the oligo(dT) hybridizes with the PolyA tail of the mRNA and mRNA is purified, the Oligo(dT) serves as a primer for the enzyme reverse transcriptase. Using the mRNA as a template, reverse trancriptase is able to synthesize a cDNA strand complementary to the mRNA (Heller et al., 2001). Therefore, mRNA purification initiates the creation of a cDNA library. The four basic steps of constructing a cDNA library are listed below.
The four basic steps in constructing a cDNA library (Miesfeld, 2000):
1. Purification of mRNA using chemical extraction and oligo-dT purification.
2. First strand cDNA synthesis using oligo-dT, random pdN6, or specific primers.
3. Second strand cDNA synthesis requires a priming event; done with RNaseH.
4. Repair of cDNA termini and ligation of adaptor oligos; clone into vector.
As compared to libraries made with other primers such as random primers and gene-specific primers, libraries created using Oligo(dT) primers have the best chance of containing long cDNAs since the priming event is targeted to the 3’ poly-A tail of the mRNA (Miesfeld, 2001).
Once you have single-stranded cDNA created, RNAse enzymes will destroy the mRNA template and DNA Polymerase I can synthesize the complementary strand. This process is outlined well in the following diagram.

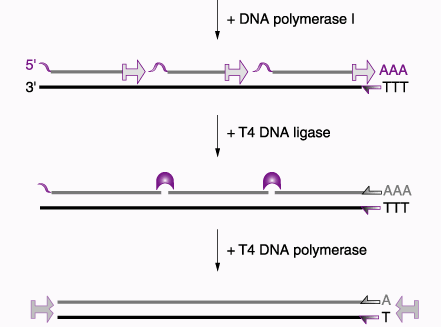
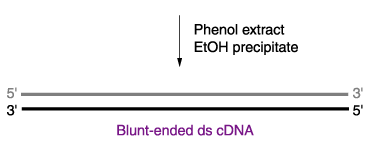
Figure 4. This figure outlines the process for creating double-stranded cDNA from purified mRNA isolated using Oligo (dT). Permission for use of this image is currently being sought from Prof. Roger Miesfeld at University of Arizona. The image is take from a lecture website: <http://www.biochem.arizona.edu/classes/bioc471/pages/Lecture9/Lecture9.html>
See next page for links to related sites.
Caroline's homepage