This web page was produced as an assignment for an undergraduate course at Davidson College.
Antisense Technology
Overview
Antisense technology is a tool that is used for the inhibition of gene expression.
The principle behind it is that an antisense nucleic acid sequence base pairs
with its complementary sense RNA strand and prevents it from being translated
into a protein. The complimentary nucleic acid sequence can be either a synthetic
oligonucleotide, often oligodeoxyribonucleotides (ODN) of less than 30 nucleotides,
or longer antisense RNA (aRNA) sequences (Sczakiel, 1997). An example of sense
and antisense RNA is: 5´ A C G U 3´ mRNA, and 3´ U G C A 5´
Antisense RNA
This type of technology was first developed by Dr. Hal Weintraub and his colleagues
at the Basic Science Division in the early 1980s. They were the first to show
that aRNA could inhibit gene expression in mouse cells (Berg, 2002). Then in
1996, Dr. Meng-Chao Yao, also at the BSD, showed how aRNA that was incorporated
into non-conserved regions of ribosomal RNA (rRNA) could also disrupt translation
(Yao, 1996) by altering the interaction of the mRNA and the rRNA/aRNA chimera.

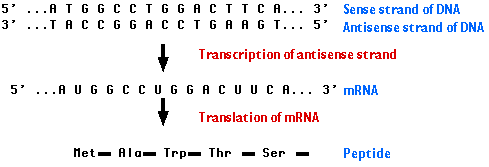
Figure1. Cartoon sequence of transcription of antisense DNA strand into the sense mRNA strand, which is then translated into a polypeptide (Kimball, Nov 2002).
Theories on how inhibition works
When the aRNA binds to the complementary mRNA, it forms a double-stranded RNA
(dsRNA) complex that is similar to double-stranded DNA. The dsRNA complex does
not allow normal translation to occur. The exact mechanism by which translation
is blocked is unknown. Several theories include:
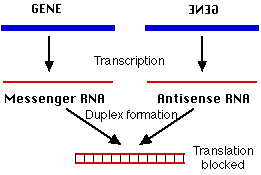
Figure 2. Cartoon of how sense mRNA and antisense RNA are trascribed and then anneal to form double-stranded RNA, which blocks translation of the protein coded for by the sense mRNA (Kimball, Nov 2002).
How to clone aRNA
In order for aRNA to block translation, it has to be inserted into the proper
cells so it can bind to its complementary sense strand. There are several ways
to incorporate aRNA into a cell. One method is to create a plasmid that codes
for the aRNA. In essence, this is a plasmid that has a promoter that initiates
transcription in the direction of the 3' end of the sense stand to the 5' end,
which corresponds to 5' to 3' transcription of the antisense strand. Next,
these liposomes can be used to transfect target cells with the newly formed
plasmids. Liposomes are cheap and efficient for this task.
Adenoviruses can also be used to infects cells and delivers aRNA sequences. This method has a higher transduction efficiency than liposomes (Tritton, 1998)
A Northern blot can be used to detect whether the aRNA is produced within the cells, and a Western blot can be used to measure amount of the mRNA gene that is expressed in wild type and mutant cells with. If the aRNA is properly expressed in the cells, then less of the mRNA gene product should be produced.
a. 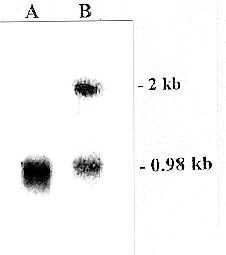 b..
b.. 
Figure 3. (a) Northern blot analysis of antisense RNA expression
in mutant (lane B) and wild type (lane A) cells. The RNA was hybridized with
in vitro labelled transcripts from the 51 kDa gene (in the sense orientation).
As a control, an in vitro labelled antisense transcript from the 21.3 kDa gene,
which encodes a subunit of the membrane arm of complex I, was included in the
hybridization. A signal with the expected 2 kb length of the antisense RNA for
the 51 kDa protein could be seen only in the transformant RNA (lane B), while
both wild type and transformant RNAs have a 0.98 kb transcript which hybridized
with the control probes for the 21.3 kDa protein. This shows that the antisense
RNA was only expressed in the wild type cells, and that the sense RNA was expressed
in both. (b)Western blot analysis of one mutant (lane
B) compared with the wild type (lane A). The same amount of mitochondrial protein
was incubated with antisera against the 51 kDa subunit and, as a control, against
the 21.3 kDa subunit of the membrane arm of complex I. The level of 51 kDa protein
in the mutant is reduced by 80% compared to the wild type. The band at 51 kDa
in lane B is significantly smaller than the band in lane A, showing that less
of the gene product was made in the cells that expressed the aRNA (Fecke, et.
al., 2003).
Problems with antisense technology
Antisense technology has not been perfected. It is still difficult to express
aRNA only in targeted tissues. Precision gene therapy using aRNA needs to be
improved because as it is right now, aRNA sometimes binds to mRNA that is not
its target. It is known to improperly bind to Human Growth Factor . Furthermore,
the uptake of aRNA is still imprecise.(Damha, 2002).
Areas of application
The possibilities that aRNA has to offer the scientific and medical fields seems
endless. Projects are in the works already on how to use antisense technology
for the deactivation of oncogenes (Kimball, May 2002), for the suppression of
viral RNA expression (Calkins, 1996), and to understand naturally occurring
aRNA that can be used for genomic imprinting (Kimball, Nov 2002).
Antisense technology has already been successful in suppressing
the gene for the protein that makes tomatoes spoil. Flavr Savr tomatoes were
transgenic tomatoes constructed to have artificial DNA that coded for aRNA that
was complementary to the RNA that coded for the protein that caused spoiling.
The aRNA suppressed the expression of this spoilage gene by 10%, which was enough
to save the tomatoes from rotting while being shipped to grocery stores. The
tomatoes are no longer on the market due to complications in the harvesting
process (Kimball, Nov 2002)
References:
Berg, Barbara. 2002 Feb 6. Antisense RNA: A mechanism to inhibit gene expression. <http://www.fhcrc.org/visitor/research/firsts/antisense.html> Accessed 2003 Feb 11.
Damha, Masad. 2002 Oct 17. Making Sense of Antisense. <http://www.erin.utoronto.ca/mbiotech/menu/damha.htm> Accessed 2003 Feb 11.
Calkins, Katrina, et. al. 1996 Dec 6. Prevention. <http://student.biology.arizona.edu/honors96/group17/dengue2.html> Accessed 2003 Feb 11.
Fecke, W., et. al. <www.fgsc.net/fgn/fecke.html> Accessed 2003 Feb 12.
Kimball, J. 2002 Nov 11. Antisense RNA. <http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/A/AntisenseRNA.html> Accessed 2003 Feb 11.
Kimball, J. 2002 May 21. BCL-2. <http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/B/BCL-2.html> Accessed 2003 Feb 18.
Sczakiel, G. 1997 Aug. The design of antisense RNA. Antisense Nucleic Acid Drug Dev. 7(4): 439-444. <http://www.aegis.com/pubs/aidsline/1998/apr/M9841776.html> Accessed 2003 Feb 11.
Tritton, T., et. al. 1998. Antisense. <www.haverford.edu/biology/HHMI/HPV/anti.html> Accessed 2003 Feb 11.
Yao Lab- Antisense RNA. 1996 Jul 3. <http://www.fhcrc.org/pubs/center_news/1996/Jul3/Antisense.htm>
Accessed 2003 Feb 11.
For questions or comments, email Heather Maloney