Low Temperature PCR

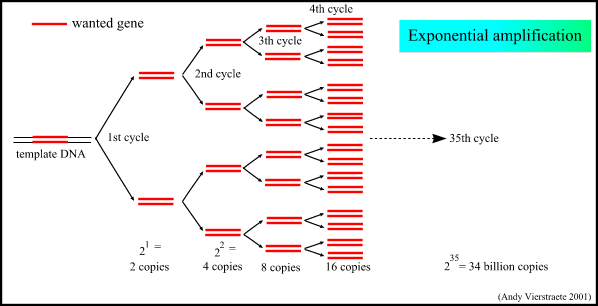
Fig. 1. This illustration shows how one template strand of DNA is amplified exponentially using the PCR process. Courtesy of http://allserv.rug.ac.be/~avierstr/principles/pcr.html (permisssion pending).
The overall concept of the polymerase chain reaction (PCR) is simple. By combining the necessary components of DNA replication in a test tube biologists can artificially replicate many copies of a given strand of DNA. The ingredients needed are template DNA, DNA polymerase, the four deoxyribonucleoside triphosphates (dATP, dCTP, dGTP, and dTTP), and primer DNA. Heating the mixture to about 95ºC denatures the template strand (HIFI Tech, 2003). Then the mixture is allowed to cool to 55ºC so the primers can bond to the known complementary sequence on the template strand. With the primers in place, DNA polymerase can bind to the template and begin replication. The entire process can be repeated by heating and cooling the mixture every few minutes.
When this process was first developed, scientists realized that the crucial heating step, which opens the double-stranded template DNA, would at the same time denature the DNA polymerase. To get around this inherent problem new DNA polymerase would have to be added for each cycle of replication, or a form of polymerase would have to be found that could withstand the extreme temperatures.
Eventually a heat-resistant DNA polymerase was isolated from Thermus aquaticus (Taq), bacteria that lives in hot springs. Since then, other forms of “thermostable” DNA polymerase have been isolated from various species and utilized in PCR reactions. “Thermostable” DNA polymerases are defined as those forms that are able to withstand temperatures of 90 to 100ºC without denaturing (HIFI Tech, 2003). The problem with these various forms of DNA polymerase is that they have low replication fidelity and low processivity due to non-specific hybridization between primers and the template.
A new form of DNA polymerase originally from Bacillus stearothermophilus (Bst) solved this problem. This form of DNA polymerase has been genetically modified, and works optimally at 70ºC. At this temperature the Bst HiFi® DNA polymerase is performs with higher fidelity than other polymerases and can replicate GC-rich areas (New England, 2003) . Under other PCR conditions, GC-rich areas are difficult to denature because the triple hydrogen bonds hold the two strands together tightly. However, the HiFi® DNA polymerase is used in conjunction with LoTemp™ PCR solution which allows these bonds to be easily broken at just 70ºC. Other components of the mixture ensure that non-specific primer hybridization is minimized (HIFI LoTemp, 2003). This form of DNA polymerase has higher replication fidelity compared to Taq polymerase and Pfu polymerase, from Pyrococcus furiosus (Viguera et. al, 2001).
Taq PCR protocol vs. Low temp Bst PCR protocol
Taq DNA polymerase 20 µl LoTemp™ PCR mixture
dNTPs and buffer mix (includes dNTPs)
two primers 1 µl each of two primers
template DNA 1 µl template DNA
deionized water 2 µl deionized water
MgCl2 (25mM)
mix above ingredients (except Taq mix above ingredients and then start
polymerase) and then start temperature cycles:
temperature cycles: 70ºC for 30 seconds
95ºC for 1-3 minutes (“hot start”) 37ºC for 20 seconds
add Taq polymerase 50ºC for 1-3 minutes
90ºC for 0.5-2 minutes repeat
75ºC for 1 minute (HIFI LoTemp, 2003)
repeat
(Fermentas, 2003)
These protocols show that the low temperature PCR procedure greatly simplifies the steps needed to amplify DNA, thus reducing cost and time spent on PCR (Mead et. al., 1991). This is coupled with cleaner results because of the nature of the Bst polymerase and the additives in the mixture. Higher fidelity results and an easier procedure make this form of PCR an excellent choice for molecular biologists who are always looking for the fastest, most reliable methods in any procedure. Visit the Hi-Fi DNA Homepage to learn more about low temperature PCR and to order supplies.
References:
Fermentas, Products for Molecular Biology. Protocol for PCR using Taq DNA Polymerase. Feb 17, 2003. Feb 18, 2003. <http://www.fermentas.com/techinfo/PCR/dnaamplprotocol.htm>
Hi-Fi DNA. Tech Services: Frequently Asked Questions. Feb 9, 2003. Feb 17, 2003. <http://www.hifidna.com/FQAall.htm>
Hi-Fi DNA. LoTemp PCR with HIFI DNA Polymerase. Feb 9, 2003. Feb 17, 2003. <http://www.hifidna.com/lowtmp.htm>
Mead, D.A., McClary, J.A., Luckey, J.A., Kostichka, A.J., Witney, F.R., and L.M. Smith. (1991). Bst DNA polymerase permits rapid sequence analysis from nanogram amounts of template. Biotechniques 11, 76-87.
New England BioLabs, Inc. Bst DNA Polymerase, Large Fragment. Jan 21, 2003. Feb 17, 2003. <http://www.neb.com/neb/products/polymerases/275.html>
Viguera, E., Canceill, D., and S.D. Ehrlich. (2001). In vitro replication slippage by DNA polymerases from thermophilic organisms. Journal of Molecular Biology 312, 323-333.
email questions and comments to: tamaloney@davidson.edu