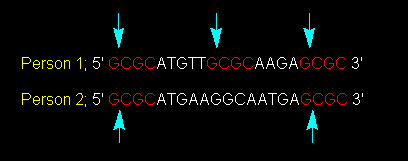*This web page was produced as an assignment for an undergraduate
course at Davidson College*
Using RFLPs for mapping genetic diseases
and for DNA fingerprinting
I. Definition:
Restriction Length Fragment Polymorphisms, RFLP's, are DNA
differences that are inherited and can be used as genetic markers for diseases
such as sickle cell anemia and phenylketonuria (Heller et al, 2001). RFLP's
can also be used to construct a family pedigree and determine paternity.
These fragments are generated by cutting genomic DNA with
a restriction endonuclease at a particular nucleotide sequence and separating
the resulting fragments on a gel by performing a Southern
blot (Campbell, 2001). The resulting RFLP's can be compared to RFLP's
from corresponding DNA sequences from family members or unrelated people.
By definition, this method detects DNA differences and therefore can only
be used to distinguish polymorphic alleles from each other.
II. Making a RFLP:
The first step in producing a RFLP is to obtain a sample
of blood, hair root, or other biological sample. The DNA from this sample
is then cut with a restriction enzyme and multiple fragments are produced
based on the DNA sequence as is shown in figure 1:
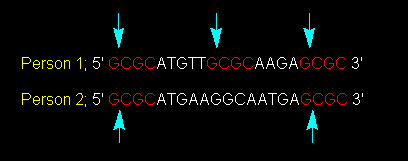
Figure 1: Cutting a Nucleotide Sequence at Particular Restriction Sites.
A particular restriction enzyme cuts genomic DNA of person 1 and 2 at the
GCGC nucleotide sequence. This image was obtained pending permission from
Dr. Simon Lewis of Deakin University and the original reference is found here.
In this figure, the blue arrows show that the restriction
enzyme cuts the nucleotide sequence between the first G and the first C. Person
1 will therefore have this DNA sequence cut into three fragments while Person
2 will have this DNA sequence cut into two fragments (Lewis, 2001). These
fragments are then run on an agarose gel in separate lanes and the fragments
will migrate towards the positive electrode to different degrees based on
the molecular weight of each fragment. (The smaller fragments will move farther
on the gel than the larger fragments.) The fragments then need to be visualized.
This is commonly done by transferring the bands to a nitrocellulose gel and
probing for the various DNA sequences contained in the fragments. Ideally,
this probe is 6-10 bp, but in the simplified example above, the probe could
be GCG, which would bind to the red CGC sequence contained by all fragments.
The probe needs to be able to be visualized, and this can be done by exposing
the radioactive probe to x-ray film. At this point, the fragments of various
lengths can be visualized and the sequence differences between these two people
can be visualized. Person one will show 3 DNA fragments and Person 2 will
show 2 DNA fragments.
III. The Use of RFLPs in Mapping Genetic Diseases
In the following illustration, DNA from the hemoglobin gene
from each family member is subjected to a particular restriction endonuclease.
Since the hemoglobin gene is polymorphic, there is more than one DNA sequence
encoding for this gene. Hb A is the wild type allele, and Hb S is the allele
that codes for the sickling of red blood cells (Huskey, 2001). RFLP's are
produced using this polymorphic DNA sequence and the resulting fragments are
separated by gel electrophoresis and as shown in Figure 2:
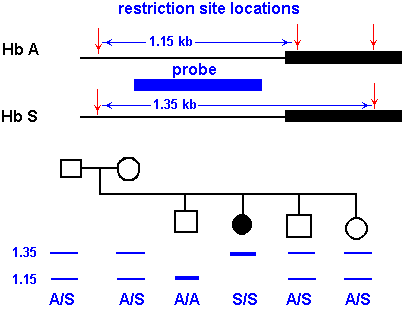
Figure 2: RFLP's produced from fragments of the hemoglobin gene. Hb A corresponds
to the wild type hemoglobin gene and Hb S corresponds to the diseased hemoglobin
gene. This image was reproduced with permission by Dr. Robert J Huskey from
the University of Virgina and can be found in its original version here.
The wild-type hemoglobin gene, Hb A, appears at 1.15 kb,
while the sickled hemoglobin gene, Hb S, appears at 1.35 kb. A person homozygous
for sickle cell anemia (S/S) shows only one RFLP at 1.35 kb, while people
heterozygous for this disease (A/S) have RFLP's at 1.35 kb and 1.15 kb. People
who have not inherited this gene (A/A) show one RFLP at 1.15 kb. (Figure 1
does not show a MW marker, but this marker is neccessary in order to determine
the MW of the fragments.)
Can we positively conclude from this one pedigree as to which
family members are homozygous and heterozygous for sickle cell anemia?
No. We must be absolutely sure that the genetic disease and
the DNA sequence that produces these RFLP's are linked. It could be that these
family members have inherited mutations on the hemoglobin gene, and that these
mutations have absolutely nothing to do with the disease. Because of this,
RFLP's need to be produced using various restriction enzymes and various DNA
sequences to be sure that the polymorphic DNA sequence is directly related
to the disease.
IV. The Use of RFLPs in DNA Fingerprinting
DNA fingerprinting is often used in criminal cases to determine
a suspect's guilt. DNA from a blood, hair root, or semen sample found at the
scene of a crime can prove guilt or innocence with high precision. The DNA sample
is cleaved with a restriction enzyme and the resulting fragments are separated
using Southern
blotting techniques. DNA from the crime scene is analyzed on the same gel
as DNA from the potential criminals, and therefore, DNA collected from the scene
can be compared with that from various suspects and the RFLP's produced from
the DNA of the guilty suspect will match with the RFLP's produced from DNA collected
from the crime scene.
In the following example, a woman was raped while she and her
fiance were sleeping in their car. They were found the next morning in the woods
next to a recreation area and both had died of gunwounds. One man was later
found driving the stolen vehicle and he told authorities of the friend that
was with him the night of the murders. In order to determine which suspect was
guilty of raping the woman, DNA fingerprinting (RFLP analysis) was used. DNA
from a semen sample retrieved from the body was compared with DNA from blood
samples from both suspects. These DNA samples were cut with a particular restriction
enzyme and fragments were separated by gel electrophoresis. Figure 3 shows the
RFLP's from the semen sample in yellow and the RFLP's from both suspects in
purple and blue. It is shown in this figure that suspect 2 is the man that raped
this woman. It is possible that an innocent person could show the exact same
restriction fragments, but the chance of this is 1 in 9,390,000,000, which is
twice the human population of the world! Suspect 2 was conviced of rape and
murder and received a double death sentence. This happened to be the first case
in the world in which the conviction of the death sentence was based on DNA
fingerprinting (The Dolan DNA Learning Center, 2000).
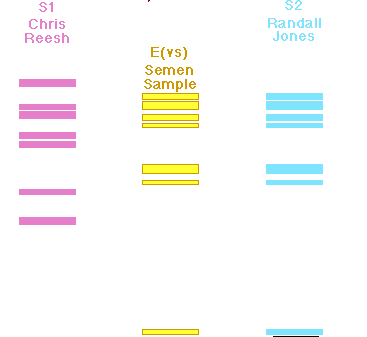
Figure 3: RFLP analysis of the semen sample collected from the
raped woman versus blood samples of two suspects. The pink and blue fragments
were produced from genomic DNA from the suspects and the yellow fragments were
produced from genomic DNA from the victim. This figure was reproduced pending
permission by the Dolan
DNA Learning Center.
References:
Campbell MA. 2001. Southern Blot Method. Davidson College, Davidson.
<http://bio.davidson.edu/Courses/genomics/method/Southernblot.html>.
Accessed 2003, February 14.
Heller HC, Orians GH, Purves WK, Sadava D. 2001. Life: the Science
of Biology, sixth edition. Sunderland, Massachusetts: Sinauer Associates, Inc.,
337.
Huskey RJ. 1996. Genotype Determination USing RFLP's and a Gene
Probe. University of Virginia. <http://www.people.virginia.edu/~rjh9u/hbsrflp.html>.
Accessed 2003 February 14.
Lewis SW. 2001 June 5. RFLP DNA Typing. Deakin University. <http://www.deakin.edu.au/forensic/Chemical%20Detective/RFLP%20DNA%20Typing.htm>.
Accessed 2003 February 17.
The Dolan DNA Learning Center. 2000. DNA Detective. <http://www.dnalc.org/resources/dnadetective.html>.
Accessed 2003, February 14.
Home
Davidson College Main Page
Molecular
Biology Main Page
Chemistry Main Page
Chemistry
Individual Research Project, May 2002
Send comments, questions, and suggestions to cawilliford@davidson.edu