This web page was produced as an assignment for an undergraduate course at Davidson College.
Melanoma Inhibitory Activity
Through analysis of the paper (Structure of melanoma inhibitory activity protein, a member of a recently identified family of secreted proteins) that first discovered the structure of melanoma inhibitory activity (MIA), I will relate the structure of the protein to its function. From past studies, the protein's name appears to be counter-intuitive with regards to its role in the cancer, melanoma. Because MIA blocks the adhesion of melanoma cells to the extracellular matrix, MIA's expression in malignant melanoma cells may contribute to the metastasis of melanoma cells. In normal, non- cancerous cells, MIA is an essential component of the cartilage cell phenotype.
Structure:
To make MIA protein, a synthetic MIA gene was cloned into a plasmid that was transformed into an Eschericia coli strain. MIA was labeled with selenomethionine, and after protein purification, MIA was extracted and visualized using multiwavelength anomalous diffraction.

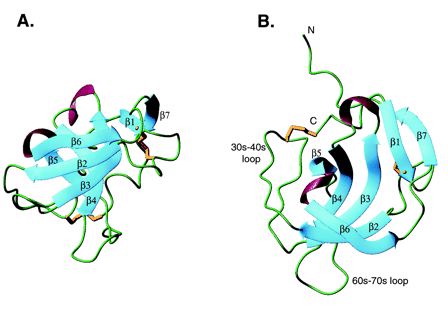
Fig. 1- Two different views of MIA. Beta sheets are in blue, alpha helices are in violet, and disulfide bonds are in gold. Green represents regions lacking secondary structure. The MIA protein structure resembles that of a barrel with (A) showing the opening of the barrel pointed to the top of the page and (B) looking into one end of the barrel. Picture courtesy http://www.pnas.org/cgi/content/full/98/10/5515/F1
The MIA protein has 7 beta sheets, 2 alpha helices, and several loops (Fig.1). MIA has a single domain and is an oblong, small globular protein with the dimensions 18X18X28 angstroms. The five beta strands (beta2-beta6) form a subdomain that is structurally similar to Src homology 3 (SH3) domains. The two antiparallel beta sheets, beta2 and beta3, form an 18-residue loop that is analogous to the RT loop found in an SH3 domain. The five strands (beta2-beta6) form a beta barrel that is flanked by the 30s-40s and 60s-70s loops (Fig.1). Adding height and widening the mouth of the barrel, MIA's N- and C- terminal extensions of the SH3 subdomain consist of 20 amino acids that wrap around the top of the barrel and build outward. The beta1 and beta7 sheets are found within these two extension sites and bond with each other and also beta sheet 3. Following beta1 is a disulfide bridge that bounds a six residue loop (Fig.1). Following beta7 are eight residues that lack any secondary structure and go from the top of the barrel to the bottom. A disulfide bridge between Cys 106 (the penultimate amino acid) and Cys35 holds these residues within the barrel structure (Lougheed 2001).
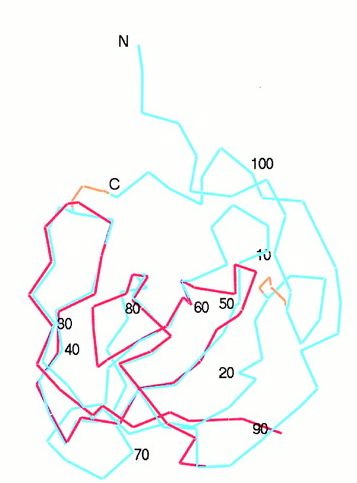
Fig.2- The SH3 domain (red) is superimposed onto the backbone of the MIA structure (blue). Picture courtesy http://www.pnas.org/content/vol98/issue10/images/large/pq0916016002.jpeg
Function:
Although MIA has an Src-homologous SH3 subdomain, MIA differs greatly in function from those proteins that contain the SH3 domain. Found in modular units of intracellular proteins, the normal SH3 domain serves to mediate protein-protein interactions through proline-rich ligands, while the secreted MIA protein has an SH3 domain created by disulfide bridges and attaches to a different class of ligands. The similarity between the RT loop of SH3 domains and the MIA 30s-40s loop may reveal more about the function of MIA as the RT loop usually denotes the ligand specifcity of the protein (Fig.1 and 2) (Lougheed 2001). Also relating to ligand specificity is the number and location of hydrophobic regions along the surface of a protein. MIA has two large hydrophobic regions along its surface. The larger of the two is found within the groove between the 60s-70s loop and the Cys-12-Cys-17 loop (Fig.3).
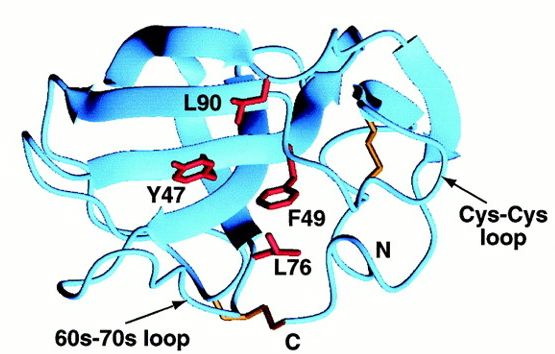
Fig.3- The larger of the two hydrophobic regions bound by the 60s-70s loop and Cys-Cys loop. Photo courtesy http://www.pnas.org/content/vol98/issue10/images/large/pq0916016003.jpeg
The hydrophobic patch consists of 9 residues, but among MIA homologues the patch is only made up of 4 residues, so the conserved area is too small to predict whether it is a ligand recognition site. The second hydrophobic patch is found between the 30s-40s loop and the first turn of alpha helix 3-10 (Fig.4).
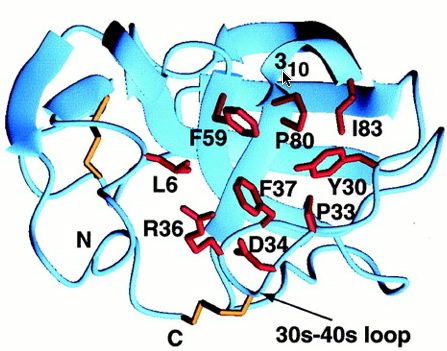
Fig.4- The second, smaller hydrophobic region bound by the 30s-40s loop and the 3subscript10 helix (cursor pointing to it). Photo courtesy http://www.pnas.org/content/vol98/issue10/images/large/pq0916016003.jpeg
Contrary to the other hydrophobic region, this one has more potential for being a site of ligand recognition. This site is the most conserved among MIA homologues, and is analogous to the polyproline helix binding site in the SH3 domain. This hydrophobic region consists of 8 residues, 4 of which are strictly conserved among MIA homologues. Although MIA appears to attach to the polyproline helix, other studies have shown that proteins with the conserved SH3 domain bind to the ligand polyproline type II (PPII). The unlikeliness of the polyproline helix binding to MIA is supported by the fact that MIA has a different structural SH3 domain than the other proteins that have this domain (Fig.5).
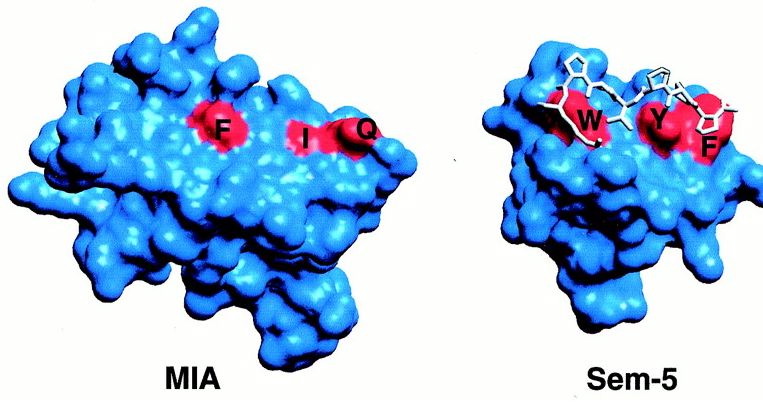
Fig.5- Comparison of MIA SH3 domain (red regioins F,I, and Q) and Sem-5 SH3 domain (red regions W, Y, and F). The polyproline helix is the white structure bound to the SH3 domain of Sem5. Photo courtesy http://www.pnas.org/content/vol98/issue10/images/large/pq0916016004.jpeg
The SH3 domain of MIA is much flatter than the domain in Sem-5 and thus, is an unlikely binding site for a polyproline helix (Fig.5). The functional diversity of MIA can be attributed to the extra fold created by the N- and C- terminal extensions (Lougheed 2001).
References:
Lougheed, J.C ., J.H., T.A., F. B., T. H. Structure of melanoma inhibitory activity protein, a member of recently identified family of secreted proteins. Proceedings of the National Academy of
Sciences of the United States of America 98:10 pgs. 5515-5520
John Bunton's Molecular Biology Home Page
Questions or Comments: email me at jobunton@davidson.edu