This web page was produced as an assignment for an undergraduate course at Davidson College.
OmpF porin Orthologs
OmpF porin
Function
To review, OmpF porin is an internal membrane protein located in the outer membrane of the Gram-negative prokaryote Escherichia coli. It functions as a non-specific ion channel that allows small, polar molecules into the cell. OmpF porin is principally utilized in cell osmotic regulation to stop or allow solutes to enter the cell and establish osmotic equilibrium between the areas internal and external to the cell. Diffusion speeds depend on the difference between the concentration outside and inside the cell (Cowan et al., 1995).
Amino Sequence
OmpF porin has an amino acid sequence that is 340 amino acids long (shown below).
1 AEIYNKDGNK_VDLYGKAVGL_HYFSKGNGEN_SYGGNGDMTY_ARLGFKGETQ
1__B_EEETTEE___ EEEEEEEEEE__ EEE__STTSSS___BSS____EE____E_EEEEEEEEEE
51 INSDLTGYGQ_WEYNFQGNNS_EGADAQTGNK_TRLAFAGLKY_ADVGSFDYGR
51 SSSSEEEEEE__ EEEEEESSS____ SSTTTTTT___E__E_EEEEEEEE__TTTEEEEEEE
101 NYGVVYDALG_YTDMLPEFGG_DTAYSDDFFV_GRVGGVATYR_NSNFFGLVDG
101 EE__ TTHHHHG_GG___ SSS_____TT___ SSSTTS__SEESSEEEEE___ ESHHHHTTTT
151 LNFAVQYLGK_NERDTARRSN_GDGVGGSISY_EYEGFGIVGA_YGAADRTNLQ
EEEEEEEE_____B__ SSSTT_ B_____ EEEEEEEE__EETTEEEEEE__ EEEEE__ HHH
201 EAQPLGNGKK_AEQWATGLKY_DANNIYLAAN_YGETRNATPI_TNKFTNTSGF
HTSSB_____SE_ EEEEEEEEEE___ EETTEEEEEE___EEEEES_ SEE_ EETTTTEEEE
251 ANKTQDVLLV_AQYQFDFGLR_PSIAYTKSKA_KDVEGIGDVD_LVNYFEVGAT
SEEEEEEEE____ EEEE__TTSEE_ EEEEEEEEEE__ ESBTTTBSEE__SEEEEEEEEE
301 YYFNKNMSTY_VDYIINQIDS_DNKLGVGSDD_TVAVGIVYQF
301 EESSSSEEEE____EEEEEE__T_TTTT______ B__ EEEEEEEEE
Figure 1. Amino acid sequence of OmpF porin. The primary amino acid of OmpF porin (shown in dark print) is 340 amino acids long. Below the primary sequence is the layout for the secondary structure of OmpF porin. As one can see, OmpF generally contains beta strands. The key is as follows: H=helix; B=residue in an isolated beta bridge; E=extended beta strand; G=310 helix; I=pi helix; T=hydrogen bonded turn; S=bend. Sequence provided by the Protein Data Bank.
Secondary Structure
The OmpF porin forms a barrel-shaped channel made out of 16 beta- helix strands. It contains 3 alpha- helix strands located throughout the structure. A long polypeptide loop named the L3 strand constricts the middle of the pore between beta strands 5 and 6. Charged residues surround this constriction and create an electrical field that slightly favors cation selectivity. OmpF porin monomers normally interact with two other monomers to form a trimer (Aboud et al, not yet published).
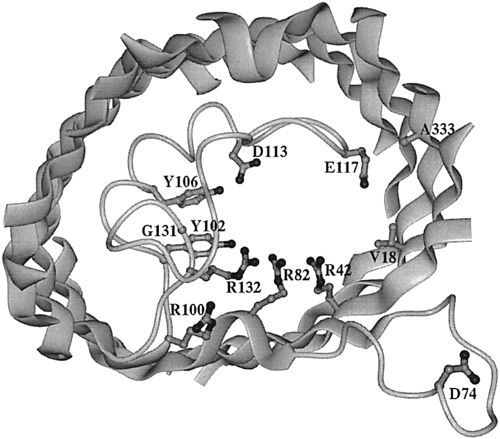
Figure 2. View of the inner OmpF porin monomer channel. The interior of the protein is the channel constriction site. R42, R82, and R132 are the positive residues within the channel. Across from them reside D113 and E117, the negative residues. The broad ribbons represent helix strands and the string represents the internal loop. The L3 polypeptide strand is located around the area of residues G131, Y102, and Y106. The D74 loop is a binding site that attaches to other OmpF porin loops in order to form a trimer (Phale et al, 2001).
Orthologs
It is important to note that OmpF porin does not have any compatible amino acid sequences with any of the eukaryotic groups of organisms. Any similarities occur solely by chance. What it does have in common with eukaryotic cells rests mainly in comparing the structure and function between the OmpF porin and the eukaryotic protein. Many of the following eukaryotic porins are of the same family and are very similar in all function and structure although their amino acids do not necessarily match. I have therefore made the assumption that if one of the eukaryotic porins is similar to OmpF porin, then it is highly likely that all the eukaryotic porins of the same family are similar to the OmpF porin.
Ortholog #1: Prokaryota, Rhodopseudomonas blastica
R. blastica is a prokaryote much like the E. coli from which OmpF comes from. R. blastula contains a channel porin that functions as a trimer within its cell membrane. Like E. coli, this porin is made of 16 anti-parallel b-helix strands that form a barrel conformation.
Within the porin, a long loop between beta strands 5 and 6 forms the pore eyelet. Like the OmpF porin, this eyelet is surrounded by electric charges with a row of positive charges on one side and a row of negative charges on the other side. This elicits an electric field that is slightly cation-selective (Kreusch et al, 1994).
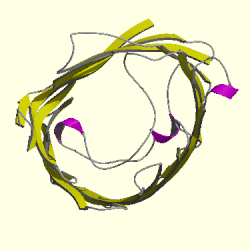
Figure 3. Cartoon of Rhodopseudomonas blastica porin. The topview of the R. blastica porin is shown. The pore eyelet is the space in the middle of the channel and curving around it is the looping polypeptide that forms this constriction. The pink ribbons represnt alpha helixes and the yellow strands that form the circle are the beta helixes. The gray strands are polypeptides that connect the beta strands to each other. Picture obtained from Cath.
Function of the R. blastica porin is also very similar to that of E. coli. The R. blastica porin is a general porin which means it allows small polar solutes of around 600 Da to passively diffuse into and out of the cell. The presence of the charged residues within its constriction space allows R. blastica porin to establish the hydrophilic environment needed for small polar molecule diffusion (Kreusch et al, 1994; Phale et al, 1994).
Ortholog #2: Eukaryota, Saccharomyces cerevisiae
S. cervisiae, more commonly known as baker's yeast, contains the outer mitochondrial membrane protein porin 1. This porin, also known as VDAC1 (voltage-dependent anion-selective channel protein 1), mainly consists of beta strands like the OmpF porin. Its 19 beta-helix strands form the "beta-barrel" structure that seems to be common among channel porins (UniProt).
VDAC1 functions as a channel that allows diffusion of small, charged molecules, like ATP and phosphate, between the cytoplasmic space within yeast cells and the inner membrane of the mitochondria. Unlike the OmpF porin, VDAC1 operates under different electrical stimuli. For example, at a low or zero membrane potential, the channel is "open" and becomes more anion-selective while at potentials 30-40 mV, it becomes "closed" and more cation-selective. (UniProt; Blachly-Dyson et al., 1997).
VDAC1 is located in the yeast's outer mitochondrial membrane. Although the OmpF porin is located on the E. coli's outer membrane and that of yeast is located on the mitochondria, this difference in location makes sense if we take into account the endosymbiont hypothesis of eukaryote cell formation. According to this theory, mitochondria (and chloroplasts) used to live separately from all other cells. At some point in time, mitochondria were either endocytosed or taken in by some other mechanism by other cells. Instead of being destroyed, mitochondria were able to adapt to living within their host; a condition that benefited both parties (Lovelock, Margulis, 1996).
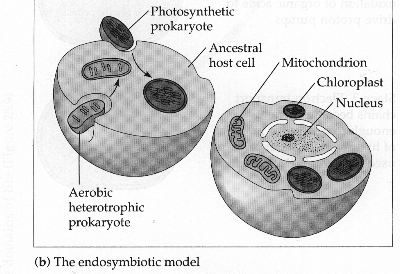
Figure 4. Illustration of the Endosymbiont hypothesis. The cell on the left demonstrates a host cell taking in an ancestral prokaryotes of mitochondria and chloroplast. The cell on the right shows the resultant effect of millions of years of mutual dependance between the host cell and these prokaryotes that have now become mitochondria and chloroplasts. Picture made by Dr. Kent Simmons.
So what signification does this have for OmpF porin and VDAC1? It provides a possible explanation for similarity between these two porins despite their non-identical amino acid sequences. Because mitochondria porin have been separated from other outer membrane porin, it is only natural that the amino acid sequences have undergone many different mutations over millions, if not billions, of years of evolution. What is important is that both structure and function between these proins has remained similar.
Ortholog #3: Eukaryota, Mus musculus
Mus musculus, the common mouse, contains the VDAC1 porin as well. Like the yeast VDAC1, this porin consists of beta-helix strands that form a channel through the outer membrane of the mitochondria.
This porin also regulates the flux of positively and negatively charged particles. Considered "open" at low membrane potentials, VDAC1 allows the transport of small anions like phosphates, chlorides, and adenine nucleotides. At high membrane potentials, VDAC1 is "closed" and allows passage of cations (Sampson et al., 1997).
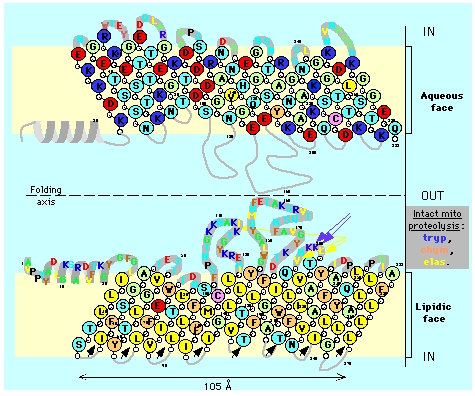
Figure 5. Topology of mitochondrial porin (VDAC). The illustration shows two views of the VDAC porin with an axis of symmetry separating them in the middle. The top porin depiction shows polar amino acids (light green), acidic amino acids (red), and basic amino acids (blue) on the aqueous face of the cell membrane. The bottom porin depiction shows hydrophobic residues (yellow) and hydrophilic residues (orange) on the lipidic face of the cell membrane. Illustration taken from Claude Aflalo.
Ortholog #4: Eukaryota, Homo sapiens
Cells from Homo sapiens also contain a VDAC1 porin on the surface of the mitochondria. Like all other VDACs, it performs the same function of passively transporting small, hydrophilic molecules. It also contains the same "beta barrel" structure (Bioinformatic Harvestor).
Other families of eukaryotic organisms that contain the VDAC protein and are therefore orthologs of OmpF porin include: Pan troglodytes, Canis familiaris, Rattus norvegicus, Gallus gallus, Drosophila melanogaster, and Caenorhabditis elegans (NCBI).
References
Cowan SW, Garavito RM, Jansonius JN, Jenkins JA, Karlsson R, Konig N, Pai EF, Pauptit RA, Rizkallah PJ, Rosenbach JP. 1995 October. The structure of OmpF porin in a tetragonal crystal form [abstract]. Structure 3(10): 1041-1050. <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=8589999>. Accessed 2005 Feb 11.
Research Collaboratory for Structural Bioinformatics (RCSB). The Protein Database. <http://www.rcsb.org/pdb/cgi/explore.cgi?job=chains&pdbId=2OMF&page=0>. Accessed 2005 March 1.
Aboud S, Marreiro D, Saraniti M, Eisenberg R. Not yet published. The role of long-range forces in porin channel conduction [abstract]. To be printed in the Journal of Computational Electronics. <http://www.iwce.nanohub.org/papers/bionano/S11-07-Aboud.pdf>. Accessed 2005 March 7.
Phale S, Philippsen A, Widmer C, Phale V, Rosenbusch J, Schirmer T. 2001 May. Role of charged residues at the OmpF porin channel constriction probed by mutagenesis and simulation. Biochemistry 40 (21): 6319-6325. <http://pubs.acs.org/cgi-bin/article.cgi/bichaw/2001/40/i21/html/bi010046k.html>. Accessed 2005 Feb 15.
Kreusch A, Neubuser A, Schiltz E, Weckesser J, Schulz GE. 1994. Structure of the membrane channel porin from Rhodopseudomonas blastica at 2.0 angstroms resolution. Protein Science 3: 58-63. <http://www.proteinscience.org/cgi/reprint/3/1/58>. Accessed 2005 March 4.
The Swiss-Prot Group. UniProt. 1987 August 5. UniProt Entry P04840. <http://au.expasy.org/cgi-bin/niceprot.pl?P04840>. Accessed 2005 March 2.
Blachly-Dyson E, Song J, Wolfgang WJ, Colombini M, Forte M. 1997 October. Multicopy suppressors of phenotypes resulting from the absence of yeast VDAC encode a VDAC-like protein. Molecular and Cellular Biology: 5727-5738. <http://mcb.asm.org/cgi/reprint/17/10/5727>. Accessed 2005 March 7.
Lovelock J, Margulis L. 1996. "The Gaia Hypothesis". Homepage. <http://www.mountainman.com.au/gaia_lyn.html>. Accessed 2005 March 8.
Sampson M, Lovell R, Craigen WJ. 1997 July. The murine voltage-dependent anion channel gene family. American Society for Biochemistry and Molecular Biology: 18966-18973. <http://www.jbc.org/cgi/content/full/272/30/18966>. Accessed 2005 March 7.
Bioinformatic Harvester. European Molecular Biology Laboratory (EMBL). <http://harvester.embl.de/harvester/P217/P21796.htm#SOURCE>. Accessed 2005 March 7.
National Center for Biotechnology Information (NCBI). <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=homologene&cmd=search&term=vdac>. Accessed 2005 March 2.
Back to Paul's Home Page
Back to Davidson College Molecular Home
Any questions, comments, or suggestions?
E-mail pachampaloux@davidson.edu



