This web page was produced as an assignment for an undergraduate course at Davidson College.
DNA Ligase

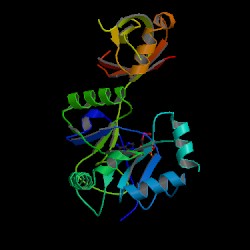
Figure 1: Chlorella virus DNA ligase-adenylate. This image is courtesy of Protein Data Bank
About DNA Ligase:
DNA ligase is an enzyme that catalyzes the repair of single-stranded phosphodiester nicks and breakages in double-stranded DNA. This protein comes in two main forms: ATP dependent and NAD+ dependent DNA ligase. Both ATP and NAD+ are cofactors that aid DNA ligase in its function. NAD+-dependent ligases have only been observed in prokaryotic organisms, while ATP-dependent ligases are found in eukaryotic, viral, and archaebacteria, (Aidan, 2000).
In vivo, DNA ligase is primarily involved in DNA replication of the lagging strand. More specifically, DNA ligase links the 3'-hydroxyl and 5'-phosphate groups of Okazaki fragments. Okazaki fragments occur as a result of the fact that the direction of replication in the lagging strand is opposite the direction in which the replication fork is moving. Therefore, in the lagging strand, DNA replication can only occur in small sections of nucleotides at a time. These small sections, or Okazaki fragments, must be ligated by DNA ligase before the DNA can be functional, (Purves, et al., 2001).
Figure 2: DNA replication fork. See this image in its original context at Cellupedia. (permission pending)
Scientists have developed many uses for DNA ligase in vitro as well. This enzyme is used as a tool to aid in the manipulation of DNA, including DNA cloning and synthesizing recombinant DNA, (Purves, et al., 2001).
Structure:
All forms of DNA ligase are comprised of at least two distinct domains. The first domain, the N-terminal domain, is the larger of the two domains and is characterized by six alpha-helices surrounding three anti parallel beta-sheets, (Aidan, 2000). This domain is also called the adenylation domain because it is the site of ATP-binding. The first domain is also believed to be the activation site for NAD+-dependent ligases. The second domain is called the C-terminus domain, or the OB fold domain. This area of the protein recognizes dsDNA. Once this region is bound to the polynucleotide, the OB fold enhances the ATP-binding of the adenylation domain of the enzyme. Because the two regions are not closely situated, the DNA ligase must undergo a conformational change in order to allow the OB domain to interact with the binding of ATP in the adenylation domain (Aidan, 2000). It is thought that throughout this conformational change, the first domain remains stationary while the second domain is free to move about and interact with the first domain. These two domains make up an enzyme complex that is essential for nucleotide ligation.
Larger DNA ligases may have additional subunits. For example, in NAD+-dependent ligases, a motif called the zinc finger is included as a subdomain of the enzyme. This subdomain aids in DNA recognition by spotting specific target sequences. In humans, it is the zinc finger in DNA ligase III that has been found to form a complex with nicked DNA. The zinc finger has also been shown to provide a structural role in ligases by supporting other subdomains, (Aidan, 2000).
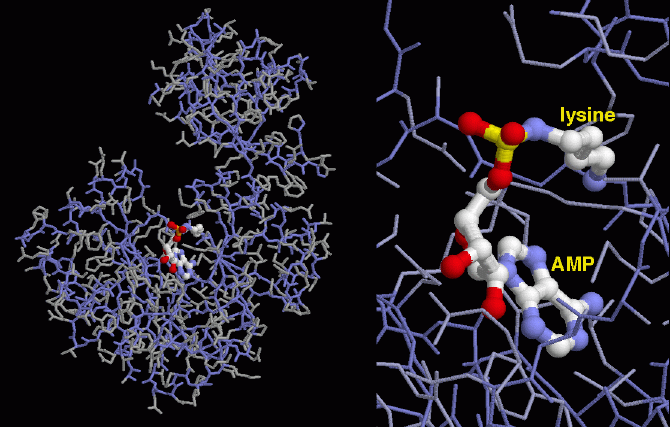
Figure 3: Model of Chlorella virus DNA ligase. Highlighted is the interaction between the activated AMP and the phosphate group of lysine in the active site. Image courtesy of Protein Data Bank.
Function:
DNA ligases use the high energy cofactors ATP and NAD+ in ligation of nicked DNA strands. The process of ligation occurs in three steps. First, DNA ligase is activated when AMP binds to a phosphate group of lysine contained in the active site of the enzyme. This AMP molecule is next transferred to the free 5' phosphate group of the nicked DNA in the second step. During the third step of ligation, DNA ligase catalyzes the attack by the 3' hydroxyl group on the 5' phosphate group, losing the AMP in the process, (Odell, et al., 2003).
Figure 4: Mechanism for ATP-dependent and NAD+-dependent ligation catalyzed by DNA ligase. This image is courtesy of Aidan, et al., 2000. You can find this image in its original context at http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=113121, (permission pending).
DNA ligases have a nick-sensing function which directs the enzyme to the correct locus of the DNA in need of repair. This function is directly correlated with the binding of the activating cofactor to the active site of the ligase molecule. If the ATP or NAD+ fail to bind to the active site of DNA ligase, the enzyme is unable to recognize the nick in dsDNA.
DNA ligase enzymes do distinguish between nicks and large gaps in DNA. It was found that a six nucleotide stretch was sufficient for DNA ligation, but any nucleotide stretch longer than this constituted a gap. In such gaps, DNA ligase effectiveness decreased drastically, (Odell, et al., 2003). It is also necessary for the nicked polynucleotide to have a 5'-phosphate group at the site of the nick. DNA ligase will not bind to a 5'-hydroxyl group.
Works Cited:
Aidan, J., Suh, S. 2000. Structural and mechanistic conservation in DNA ligases. Nucleic Acids Research 28(21): 4051-58
Cellupedia. Cheng, D., Wang, R., Zhang, F., Silbermann, B. Cellular Processes: DNA Replication. http://library.thinkquest.org/C004535/dna_replication.html?tqskip1=1 Accessed 2005 Feb 12.
Odell, M., Malinina, L., Sriskanda, V., Teplova, M., Schuman, S. 2003. Analysis of the DNA joining repertoire of Chlorella virus DNA ligase and a new crystal structure of the ligase-adenylate intermediate. Nucleic Acids Research 31(17):5090-100
[PDB]Protein Data Bank. Goodsell, D. Molecule of the Month: DNA ligase. http://www.rcsb.org/pdb/molecules/pdb55_3.html Accessed 2005 Feb 12.
[PDB]Protein Data Bank ID 1P8L. Odell, M., Malinina, L., Sriskanda, V., Teplova, M., Shuman, S.: Analysis of the DNA Joining Repertoire of Chlorella Virus DNA Ligase and a New Crystal Structure of the Ligase-Adenylate Intermediate Nucleic Acids Res. 31 pp. 5090 (2003)http://www.rcsb.org/pdb/cgi/explore.cgi?job=graphics&pdbId=1FVI&page=0&pid=165511108240335
Purves, W., Sadava, D., Orians, G., Heller, H. 2001. Life, Sixth Edition. Massachusetts: Sinauer Associates, Inc. 1044 p.
Davidson College Biology Home Page
© Copyright 2005 Department of Biology, Davidson College, Davidson, NC 28036
If you have any questions, comments, or suggestions concerning this page, please contact sadurnbaugh@davidson.edu