(this site was created as an assignment for Molecular Biology, an undergraduate course at Davidson College)
MY FAVORITE PROTEIN:
Prostaglandin H2 synthase 1 (PGHS)
What is PGHS? What does it do?
PGHS is a monotopic membrane protein found chiefly in the endoplasmic reticulum in nearly all tissues. It has an ellipsoid shape comprised of two of the same monomers.
It plays a major role in the arachidonate cascade as a bifunctional enzyme:
~cyclooxygenase reaction: converts arachidonic acid ---> prostaglandin G2 (PGG2).
~peroxidase reaction: converts prostaglandin G2 ---> prostaglandin H2 (PGH2).
The catalysis of arachidonic acid to PGH2 by PGHS is the first step in the production of prostaglandins.
(Picot, Loll and Garavito, 243)
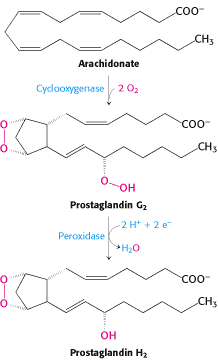
Figure 1. Formation of Prostaglandin H2. In two steps, the bifunctional enzyme PGHS catalyzes arachidonic acid to prostaglandin H2. (W.H. Freeman and Company, 2002)
What is the structure of (ovine) PGHS?
Primary Structure
600 amino acids (with a 24 amino acid signal sequence)
MSRQSISLRFPLLLLLLSPSPVFSADPGAPAPVNPCCYYPCQHQ
GICVRFGLDRYQCDCTRTAIPAPTAPSRRYGPGSGRLCGPAPLSSTFMLTHGRWLWDF
VNATFIRDTLMRLVLTVRSNLIPSPPTYNIAHDYISWESFSNVSYYTRILPSVPRDCP
TPMGTKGKKQLPDAEFLSRRFLLRRKFIPDPQGTNLMFAFFAQHFTHQFFKTSGKMGP
GFTKALGHGVDLGHIYGDNLERQYQLRLFKDGKLKYQMLNGEVYPPSVEEAPVLMHYP
RGIPPQSQMAVGQEVFGLLPGLMLYATIWLREHNRVCDLLKAEHPTWGDEQLFQTARL
ILIGETIKIVIEEYVQQLSGYFLQLKFDPELLFGAQFQYRNRIAMEFNQLYHWHPLMP
DSFRVGPQDYSYEQFLFNTSMLVDYGVEALVDAFSRQPAGRIGGGRNIDHHILHVAVD
VIKESRVLRLQPFNEYRKRFGMKPYTSFQELTGEKEMAAELEELYGDIDALEFYPGLL
LEKCHPNSIFGESMIEMGAPFSLKGLLGNPICSPEYWKASTFGGEVGFNLVKTATLKK
LVCLNTKTCPYVSFHVPDPRQEDRPGVERPPTEL (DeWitt and Smith, 1988)
Secondary Structure
Predominantly helical constitution, almost no beta-pleated sheets
(Picot, Loll and Garavito, 243)
Tertiary Structure
Three Folding Units
-
Epithelial Growth Factor-like domain
-
Membrane Binding Motif
-
Catalytic Domain
(Picot, Loll and Garavito, 243)
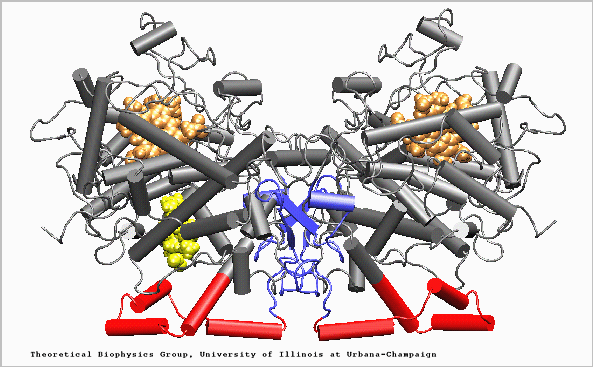
Figure 2. Structure of the Prostaglandin H2 synthase-1 homo-dimer. EGF-like domain (blue), membrane binding motif (red), catalytic globular domain (grey), heme groups (brown), and arachidonic acid in its binding site (yellow) (Molnar, 1999) permission pending
FOLDING UNIT #1 - EGF-like domain
amino acids 34-72
small and compact
two two-stranded beta pleated sheets held together by three disulfide bonds
connected to the main body by a disulfide bond b/t Cys37 and Cys157
found b/t the monomers just outside the membrane binding motif
(Picot, Loll and Garavito, 249)
FOLDING UNIT #2 - Membrane Binding Motif
amino acids 73-116
4 amphipathic helicies (A-D), allowing insertion into the bilayer
inserted into one leaflet of the bilayer
transitional helix D connects this unit to the main body
(Picot, Loll and Garavito, 247-248)
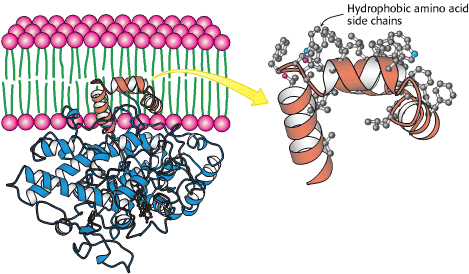
Figure 3. Attachment of Prostaglandin H2 Synthase-1 to the Membrane. α helices with hydrophobic side chains hold PGHS in one leaflet of the membrane. (One monomer of the dimeric enzyme is shown.) (W.H. Freeman and Company, 2002)
FOLDING UNIT #3 - Catalytic Domain
-large, globular
-two distinct chains interwoven chains connected by six segments of polypeptide chain
-
large lobe
V shaped
3 core helicies surrounded by 4 helicies and intervening loops
-
small lobe
bundle of 6 nearly parallel helicies
(Picot, Loll and Garavito, 243)
-contains the adjacent cyclooxygenase & peroxidase active sites
-
peroxidase active site
haem-dependent
found b/t large and small lobes in a shallow groove
(Picot, Loll and Garavito, 244)
-
cyclooxygenase active site
hydrophobic long, narrow channel
found on the outer surface of the membrane binding motif in the middle of helicies A, B and C to the center of the monomer
(Picot, Loll and Garavito, 245-246)
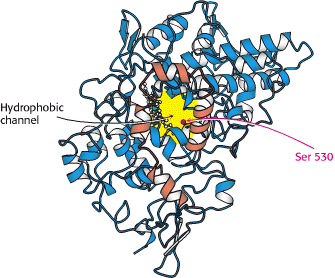
Figure 4. Hydrophobic Cyclooxygenase Channel of Prostaglandin H2 Synthase 1. A view of prostaglandin H2 synthase from inside of the membrane, showing the hydrophobic channel that leads to the cyclooxygenase active site. The helices bound to the membrane are shown in orange. (W.H. Freeman and Company, 2002)
How does structure affect function?
The cyclooxgenase site contains Ser 530 which is acetylated by aspirin. Ser 530 is positioned where its acetylation blocks access of arachidonic acid to the active site (Picot, Loll and Garavito, 246).
Hydrophobic groups on specific sides of alpha helicies allow connection of the membrane binding motif which leads to the hydrophobic channel and the cyclooxygenase active site. This allows molecules like fatty acids to directly reach the active site from inside the bilayer.
The enzyme is structured so that the active sites are adjacent allowing the product of the first reaction, PGG2 to move directly to the peroxiadase site.
References
Berg JM, Tymoczko JL, Stryer L. 2002. Biochemistry 5th Ed. <http://www.ncbi.nlm.nih.gov/books/bv.fcgi?call=bv.View.ShowTOC&rid=stryer.TOC&depth=10>
DeWitt DL, Smith WL. 1988. Primary structure of prostaglandin G/H synthase from sheep vesicular gland determined from the complementary DNA sequence. Proceedings of the National Academy of Sciences of the United States of America 85(5). <http://www.pubmedcentral.nih.gov/pagerender.fcgi?artid=279781&pageindex=1#page>
Molnar F. 1999 Sep 13. Theoretical and Computational Biophysics Group Prostaglandin H2 synthase-1 Research page. <http://www.ks.uiuc.edu/Research/pghs>
Ophardt CE. 2003. Virtual ChemBook. <http://www.elmhurst.edu/~chm/vchembook/555prostagland.html>
Picot D, Loll PJ, Garavito RM. 1994. The X-ray crystal structure of the membrane protein prostaglandin H2 synthase-1. Nature 367:243-49.
please email me with any questions or comments
access the Molecular Biology homepage