This web page was produced as an assignment for an undergraduate course at Davidson College
Website Navigation:
Spike Protein Structure and Function
More Information on SARS and the Spike Protein:
Centers for Disease Control and Prevention
Other Interesting Viral Proteins:
Webpage created by Kevin Saunders, an undergraduate student at Davidson College.
The carboxyl terminus (C-terminus) is comprised of the transmembrane region and the cytoplasmic tail (Rota et al., 2003). The tail is cysteine rich and highly conserved among all variations of the SARS-CoV (Rota et al., 2003). The transmembrane region and cytoplasmic tail are only 52 amino acids long, which is the shortest length of all known coronavirus S proteins (Rota et al., 2003).
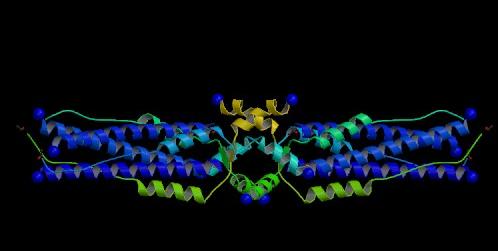
The extracellular domain of the Spike protein is comprised of two heptad repeat regions. These regions are called heptad repeat region 1 (HR1) and heptad repeat region 2 (HR2), or HR-N and HR-C to denote the closest terminus to the HR (Rota et al., 2003; Xu et al., 2004).
Figure 1. The S2 subunit of the Spike (S) protein of SARS-CoV. A ribbon representation of the S2 subunit of the S glycoprotein. This subunit is responsible for cellular and viral membrane fusion.
Image courtesy of PDB
HR1 and HR2 are separated by approximately 140 amino acids called the interhelical domain (Tripet et al., 2004). The two HRs can form a six helix bundle comprised of three helices from HR1 that run antiparallel to three helices from HR2 (Xu et al., 2004). Collectively, all six helices are called the fusion core (Xu et al., 2004).
In some strains of SARS-CoV the S protein is broken into two non-covalently bonded subunits (S1 and S2) (Tripet et al., 2004). The mechanism for how the S protein is broken into S1 and S2 is still debated. The amino acid sequence does not suggest that the S protein is cleaved into two subunits, but cleavage is the primary mechanism used by other coronaviruses (Rota et al., 2003; Tripet et al., 2004).
Function
Spike (S) proteins of coronaviruses are type I membrane glycoprotein located in the viral envelope (Rota et al., 2003). These glycoproteins mediate binding of viral particles to cells, as well as cell-viral membrane fusion (Rota et al., 2003).
The S protein binds to a receptor on the host cell (Rota et al., 2003). The receptor binding is between residues 303 and 537 (Xiao et al., 2003). The S1 subunit contains these residues, thus it controls what cells can be targeted for infection by SARS-CoV (Tripet et al., 2004).
S2 is the transmembrane subunit that facilitates viral and cellular membrane fusion (Tripet et al., 2004). Membrane fusion occurs when there is a conformational change in the HRs to form a fusion core. The HRs of the protein fold into coiled-coil structure-called the fusogenic state-causing the HR domains of the S protein to fold into a hairpin-like formation (Tripet et al., 2004). This hairpin structure results in the cellular and viral membranes being pulled together and ultimately fusing (Tripet et al., 2004; Rota et al., 2003).
Image courtesy of PDB
Rota, P. et al. 2003. Characterisation of a novel Coronavirus associated with Severe Acute Respiratory Syndrome. Science 300: 1394-1399.
Tripet, B., Howard, M., Jobling, M., Holmes, R., Holmes, K., and Hodges, R. 2004. Structural characterization of the SARS-Coronavirus Spike S fusion protein core. The Journal of Biological Chemistry 279(20): 20836-20849.
Xiao, X. Chakraborti, S., Dimitrov, A., Granatikoff, K., and Dimitrov, D. 2003. The SARS-CoV S glycoprotein: expression and functional characterization. Biochemical aand Biophysical Research Communications 312: 1159-1164.
Xu, Y., Lou, Z., Liu, Y. Pang, H., Tien, P., Gao, G., adn Rao, Z. 2004. Crystal structure of Severe Acute Respiratory Syndrome Coronavirus Spike protein fusion core. The Journal of Biological Chemistry 279 (47): 49414-49419.