Davidson College Molecular Biology
Transposase

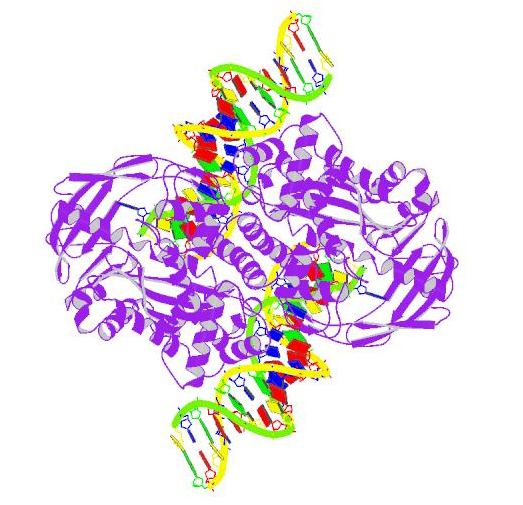
Crystal Structure of Tn5 Transposase Complexed With Transposon End DNA (Protein Data Bank).
Background

Barbara McClintock In Lab With Maize (National Library of Medicine).
In the late 1940s and early 1950s Barbara McClintock conducted several studies on the mosaic kernel colors of maize. She observed the unstable patterns of inheritance and concluded they were the result of mobile sections of the genome. She noted two loci, Dissociator (Ds) and Activator (Ac), were the cause of the mobility and resulting unstable phenotypes. (McClintock, 1952) Thirty years later her discovery of transposition won her the Nobel Prize for Medicine. Transposition is the process by which a section of a chromosome (donor DNA) is excised and reinserted in a different chromosomal (target DNA) location. The transposable element is known as the transposon. DNA transposons exist in multiple varieties including Tn3, Tn5, Tn7, and Tn10. A generic transposon contains highly specific DNA sequences at each end (these mark the transposon boundaries), a gene or multiple genes of interest such as antibiotic resistance, and a gene encoding the enzyme transposase. The enzyme, transposase, carries out the transposition. It recognizes the marking sequences, binds, excises the transposon, and inserts it into a new location (Davies et al., 2000). The importance of understanding transposase holds weight for HIV research. Integrase, the protein used by retroviruses to insert viral DNA into host DNA, functions similarly to transposase, perhaps due to a nearly identical active site. By understanding the manner in which transposase functions, further insights can be made into integrase's mechanisms thereby creating the opportunity for better anti-HIV drugs.
Transposon Structure
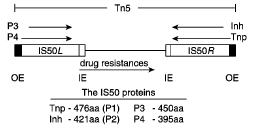
Figure 1. The Tn5 Transposon. Note the arrangement of the outside end (OE) and inside end (IE) sequences around the protein encoding genes (Goryshin et al., 1998).
The Tn5 transposon (Figure 1) contains two sets of 19bp binding sequences and three genes for antibiotic resistance between two IS50 elements. IS50R and IS50L each contain transposase (Tnp) and Inh, but IS50L is inactive (Goryshin et al., 1998). Inh inhibits transposition by forming an oligomer with Tnp thereby preventing it from properly carrying out its functions. This inhibitor is a naturally occurring deletion variant of Tnp and lacks 55 amino acids in its N-terminal end (de la Cruz et al., 1993). The two sets of biding sequences consist of two identical outside end (OE) sequences and two identical inside end (IE) sequences. The IE sequence is 5’CTGTCTCTTGATCAGATCT3’ and the OEs sequence is 5’CTGACTCTTATACACAAGT3’ (Bhasin et al., 1999). Tnp can bind to either of these sequences, but in order to transpose the entire transposon it must bind to both OE sequences. To transpose just the Tnp and Inh encoding region (IS50R), Tnp binds to an OE and an IE.
Mechanism
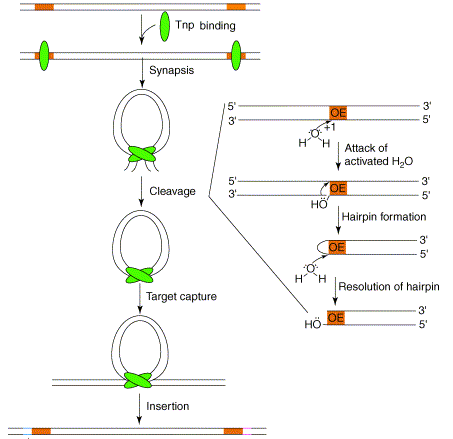
Figure 2. Cartoon overview of Tn5 transposition. The orange boxes are the outisde end sequences. The green oblongs are the monomers of Tnp. Note the formation and resolution of the hairpin through hydrolysis (Steiniger-White et al., 2004).
Tn5 transposase performs a “cut and paste” transposition meaning the donor DNA is first fully excised from its original chromosomal location and then inserted into a new location. Tnp is a multifunctional protein with its functions controlled by allosteric interactions. Its functionality also depends on intra-DNA events. Transposition begins when the two monomers of Tnp recognize and bind to the target sequences. The monomers then homodimerize through their C-termini to form a synaptic complex. Simultaneously at both ends of the transposon (Figure 2), Tnp cuts the 3’ end of the OE (or an IE) through hydrolysis exposing a 3’OH. In other words the phosphodiester backbone is nicked. The exposed 3’OH then attacks the 5’ end on the opposite strand thus forming a hairpin intermediate at each end. At this point the transposon and its synaptic complex are completely free from the donor DNA. The hairpin intermediate is resolved through hydrolysis, which regenerates the 3’OH for insertion into the target DNA. Tnp then captures a target DNA molecule and the 3’OH executes a nucleophilic attack, a transesterification reaction, on the target strand to insert the transposon (Steiniger-White et al., 2004). The beauty of this reaction is that it allows for Tnp to be the only enzyme involved by having the DNA participate in the nicking. There does not seem to be a specific DNA sequence required for Tnp binding and insertion, however Tnp does exhibit a preference. Its preferred DNA target sequence is A-GNT(T/C)(A/T)(A/G)ANC-T where N is any nucleotide and nucleotide/nucleotide is one or the other (Goryshin et al., 1998, Tn5/IS50).
Transposase Structure
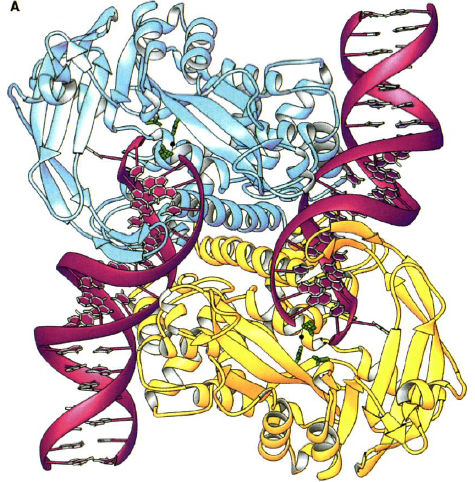
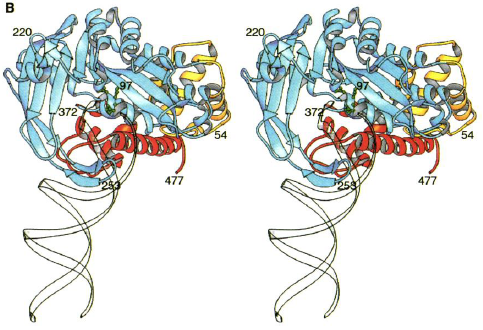
Figure 3.( A) Structure of Tn5 Tnp complex. The blue is one monomer of Tnp and the yellow is the other. The DNA is in red. The green are the catalytic active site residues.
(B) Individual Tn5 Tnp monomer. The yellow is the N-terminal domain. The blue is the catalytic domain. The red is the C-terminal domain. The green are the active site residues. The DNA is the white ribbon (Davies et al., 2000).
Each identical monomer of transposase (Tnp) contains 476 amino acids, which are divided into three major folding domains: a 70-residue N-terminal domain, a 100-residue C-terminal domain, and a 300-residue catalytic domain between (Figure 3B). The secondary structure of the N-terminal domain contains only &alpha-helices and turns and primarily binds to DNA (known as cis binding). It has four &alpha-helical regions one of which functions as the major DNA recognition helix. The major amino acid players for cis binding are glutamate and three arginines. The C-terminal domain also contains only &alpha-helices and turns, but primarily facilitates protein-protein interactions between the Tnp monomers. The catalytic domain contains an &alpha&beta&alpha fold motif. The &beta sheet is composed of five strands with the second strand anti-parallel to the others. The catalytic domain participates in an extra DNA binding by way of a second &beta sheet, which exists just after the fifth &beta strand and before the &alpha helix. This 90-residue sequence contains four anti-parallel strands of which two are long enough to reach beyond the active site to bind (known as trans binding) to the DNA bound by the N-terminal domain of the other monomer. Thus, each monomer is additionally bound to the DNA catalyzed by the other monomer.
The catalytic domain contains the active site of Tnp. There is just one active site in each monomer. It contains the DDE (aspartic acid and glutamic acid) amino acid motif. Just like other transposases, Tn5 Tnp uses a divalent metal ion for catalysis. However, instead of the common two Mg2+, Tn5 Tnp uses one Mn2+. The Mn2+ is coordinated by an aspartic acid and glutamic acid and is involved in the regeneration of the 3’OH for nucleophilic attack on target DNA. Since this model contains a hairpin formation, the 3’OH must be close enough to the 5’ phosphate group for transesterification to occur. It is only 3.5 Angstrom units away, which is accommodative. Also accommodative to the 3’OH is a trench containing multiple basic amino acid side chains. This trench is wide enough for DNA to fit and allow for a nucleophilic attack (Davies et al., 2000).
While the monomer has separate structural domains, it does not have completely separate functional domains. The previous paragraphs describe primary functions, but all three domains participate in both DNA binding and formation of the synaptic complex. The formation of the synaptic complex is a key regulatory event. The homodimerization prevents just one OE from being excised and causes coordinated cleavage and insertion of the transposon .
1. Bhasin, A, Goryshin,IY, Reznikoff, WS. 1999 December. Hairpin formation in Tn5 Transposition. The Journal of Biological Chemistry; 274 (52): 37021-37029.
2. Davies, DR, Goryshin, IY, Reznikoff, WS, Rayment, I. 2000 July. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science; 289 (5476): 77-85.
3. de la Cruz, NB, Weinreich, MD, Wiegand, TW, Krebs MP, Reznikoff, WS. 1993 November. Characterization of the Tn5 transposase and inhibitor proteins: a model for the inhibition of transposition. Journal of Bacteriology; 175 (21): 6932-6938.
4. Goryshin, IY, Miller, JA, Kil YV, Lanzov, VA, Reznikoff, WS. 1998. Tn5/IS50 target recognition. Proceedings of the National Academy of Sciences 95: 10716-10721.
5. Goryshin, IY, Reznikoff, WS. 1998. Tn5 in vitro transposition. The Journal of Biological Chemistry 273: 7367-7374.
6. McClintock, B. 1952. Mutable loci in maize. Carnegie Institute of Washington Yearbook 51: 212-219.
7. Steiniger-White, M, Rayment, I, Reznikoff, WS. 2004 February. Structure/function insights into Tn5 transposition. Current Opinion in Structural Biology; 14 (1): 50-57.
Related Readings
Molecule of the Month: Transposase
Jmol Structure of Tn5 Transposase
Structure of Prototype Foamy Virus: A Model for HIV-1 Integrase
Davidson College Molecular Biology
Questions? Comments? E-mail Bobby DesPain