Human Pepsin A
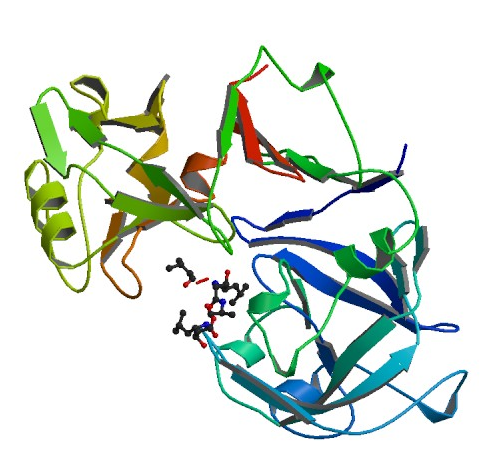
Figure 1. Pepsin complexed around pepstatin
Source: Protein Data Bank
*This website was produced as an assignment for an undergraduate course at Davidson College.*
Human Pepsin A

Figure 1. Pepsin complexed around pepstatin
Source: Protein Data Bank
Pepsin terms a small group of gastric proteases that are active in acidic environments with a pH between 1 and 5. Its name comes from the Greek word pepsis, which means to digest. The most studied and commercially available form of pepsin is porcine pepsin A, isolated from the gastric mucosa of a pig. Pepsin is not directly formed after translation of its coding mRNA, but instead begins as a zymogen, or an inactive precursor. This preliminary, inactive form that is initially translated is called pepsinogen. The activation of pepsinogen is accomplished by lowering the pH below 4.5, which leads to a cascade of changes in bond structure, as shown in Figure 2 below, and yields the enzyme pepsin. The first step is reversable, however once the protein has progressed beyond step II, the protein cannot revert back to the inactive pepsinogen. (James and Sielecki, 1986)
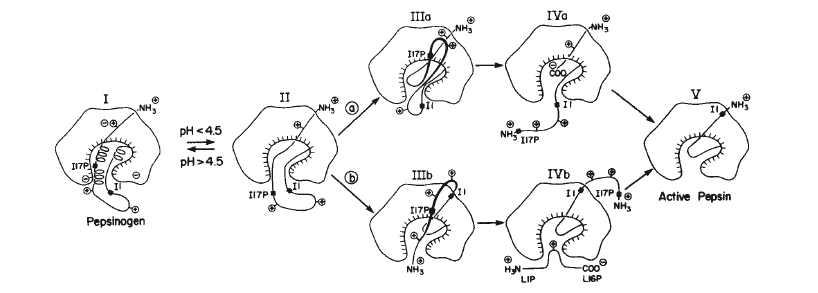
Figure 2.
Proposed steps in the activation of pepsinogen into pepsin. Note that pepsin ends up with 44 amino acids less than pepsinogen, left out of the final enzyme after step IV.
Source: James and Sielecki, 1986
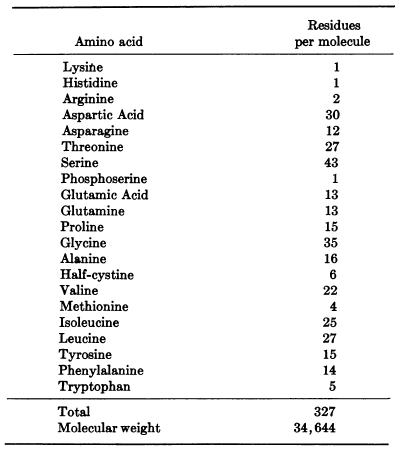
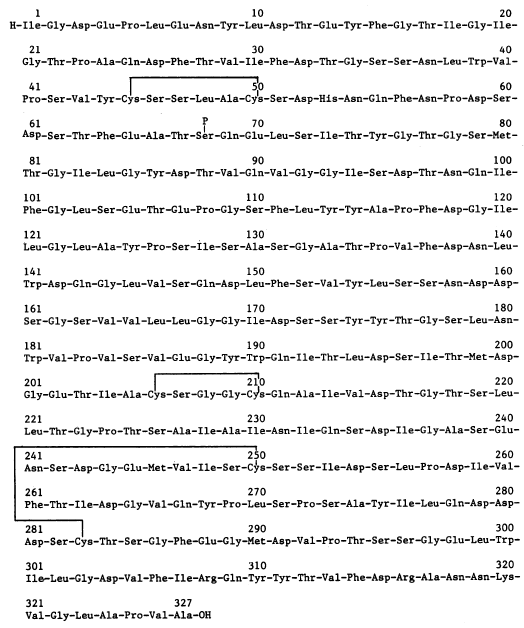
Figure 3. Amino acid sequence and overall makeup of porcine pepsin A
Source: Tang, et al. 1973
Porcine Pepsin A has a molecular weight of 36,000 Da and is made up of 327 amino acids. The protein is made up of two domains with intracellular symmetry, as observed in other aspartyl proteases. It has been proposed by that this structure is due to a duplication of a gene corresponding to a pair of identical precursor proteins that fused to form the pepsin we find today. Support for this theory has come from the finding that the aspartic proteinases of the human immunodeficiency virus (HIV-1) and the Rous sarcoma virus (RSV) are dimeric proteins in which two separate subunits correspond to the lobes of pepsin. (Pearl and Taylor, 1987)
Pepsinogen, the inactive protein that transforms to pepsin at low pH, has an additional 44 amino acids on its N-terminus that are released during the transformation. All aspartyl proteases belong to the class of "β-proteins". As the name reveals, pepsin is made up mostly of β-sheets with only 6 observed helical sections, none consisting of more than 10 amino acids. Pepsin has fewer basic amino acid residues than any other proteins as shown in figure 3: 1 lysine, 2 arginines, and 1 histidine. In contrast, the enzyme has 44 acidic residues. This helps explain pepsin's stability at extremely low pH because positive charges in acid media decrease the stability of polymeric structures. The complex tertiary hydrogen bonding of the molecule between the β sheets and other elements further contributes to the structure's acidic stability. Pepsin also has 3 disulfide bridges. Pepsin, as depicted in figures 1 and 4, has a crescent moon shape with a large, obvious active site. This site is inhibited in Figure 1 by the potent protease inhibitor pepstatin. (Andreeva, et al. 1893)
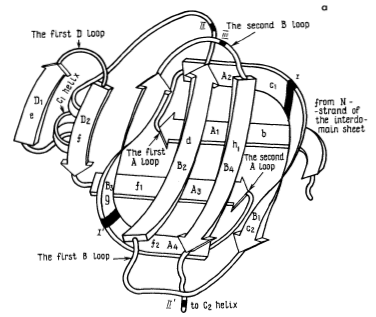
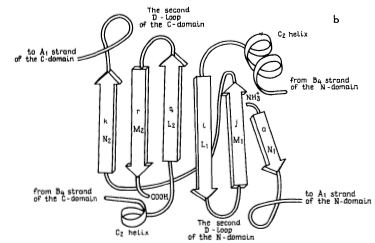
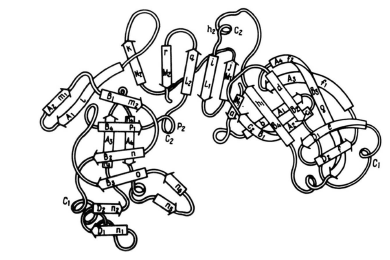
Figure 4. These three images model the tertiary structure of porcine pepsin A. The leftmost image, labeled a, depicts the intradomain double-layer mixed β-sheet that can be found in each of the two domains. The center image, labeled b, depicts the interdomain section mostly consisting of β-sheets. These two elements together can be seen in the third image making up the protein as a whole. The elements from two domains with the complex double-layer structure can be seen on either side of the interdomain section.
Source: Andreeva et al. 1983
Pepsin is an enzyme which breaks down polypeptides through a general acid-base catalysis in which water is an essential participant. This process involves the abstraction of a protein from water, so the low pH atmosphere plays a central role in the enzyme's function. The pH causes the denaturation of most proteins, ensuring the tertiary structure of these polypeptides does not prevent the active site of pepsin from breaking them down. Porcine pepsin A is found in the gut of pigs and a very similar pepsin is also present in the human gut. This pepsin is released by the gut following the ingestion of food by the organism so that the proteins in the food can be broken down and eventually turned into energy. The signal pathway is begun by the vagus nerve and leads to the release of both gastric acid, also known as hydrochloric acid, and pepsinogen. The hydrochloric acid lowers the pH, triggering the conversion of inactive pepsinogen into active pepsin and facilitating the breakdown of any polypeptides in the ingested food. (Fruton, 2002)
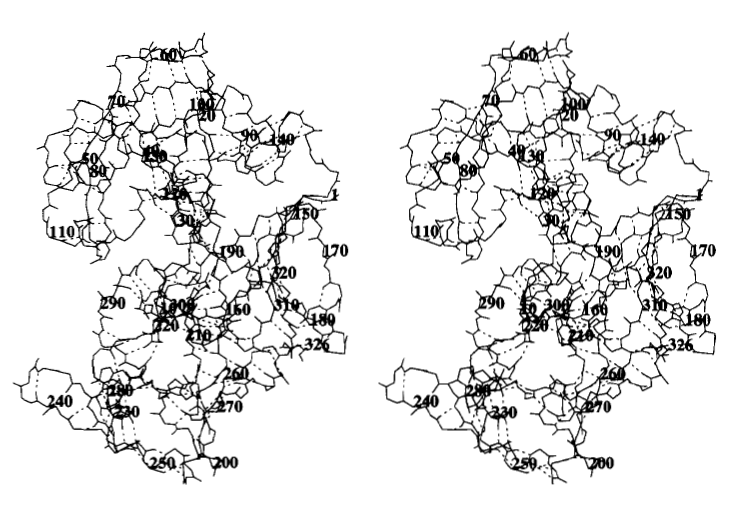
Figure 5. Stereogram showing the overall structure of human pepsin from both sides. Main-chain atoms are shown with the associated hydrogen bonds shown with broken lines. Note that this is an image of human pepsina nd may differ slightly from the previous Figures of porcine pepsin A.
Source: Fujinaga et al. 1995
1. James M, Sielecki A. Molecular structure of an aspartic proteinase zymogen, porcine pepsinogen, at 1.8 Angstrom resolution. Nature. 1986; 319: 33-38
2. Tang J, Sepulveda P, Jarciniszyn, Jr J, Chen K, Huang W-Y, Tao N, Liu D, Lanier J. Amino-Acid Sequence of Porcine Pepsin. Proc Nat Acad Sci 1973; 70, 12, I: 3437-3439.
3. Pearl L, Taylor W. A structural model for the retroviral proteases. Nature 1987; 329: 351-354.
4. Andreeva N, Zdanov A, Gustchina A, Fedorov A. Structure of Ethanol-inhibited Porcine Pepsin at 2-Angstrom Resolution and Binding of the Methyl Ester of Phenylanyl-diiodotyrosine to the Enzyme. Journal of Biological Chemistry 1983; 259, 18: 11353-11365.
5. Fruton J. A History of Pepsin and Related Enzymes. The Quarterly Review of Biology. 2002; 77, 2: 127-143.
6. Fujinaga M, Chernaia M, Tarasova N, Mosimann S, James M. Crystal Structure of human pepsin and its complex with pepstatin. Protein Science,1995; 4: 960-972.
Please direct questions or comments to algonzalezstewart@davidson.edu.