This web page was produced as an assignment for an undergraduate course at Davidson College.

Background:
Kinases are enzymes that transfer phosphate groups from high energy molecules to substrate groups. Protein Kinase C (PKC), a specific type of kinase enzyme, is composed of a family of single polypeptide enzymes that help mediate signal transduction cascades by hydrolyzing lipids. Most notably, PKC plays a role in: modulating membrane structure, regulating transcription, mediating immune responses, regulating cell growth, and is linked with receptor desensitization (Newton, 1995). In the mid 1980’s, research on PKC discovered that this enzyme consisted of a subgroup of enzymes differing by their expression of four conserved domains: C1-C4. These different forms of PKC would be distinguished by their structure and cofactor regulation into three subgroups: conventional, novel, and atypical. In order to achieve optimal activty, all isozymes require the acidic lipid phosphatidylerines, which exclusively lies on the cytoplasmic side of membranes. Of these isozymes, both cPKC and nPKC utilize protein kinase C's fundamental second messenger and activator diacylglycerol, DAG (Newton, 1995). While other factors like Ca2+ and Phorbol esters are sufficient for PKC activation, this varies between isozyme sturcture. This activation is traditionally determined by PKC isoform translocation seen through immunoblotting of subcellular segments (Schmitz-Peiffer and Biden, 2008). Protein kinase C and many of its discoveries were based off of the archetypal kinase, protein kinase A. Unlike PKA, protein kinase C has ATPase and phosphatase activity, displays lower stereospecificity allowing PKC to bind either C to N or N to C terminus cites, and PKC autophosphorylates in vitro (Newton, 1995).
Structure:
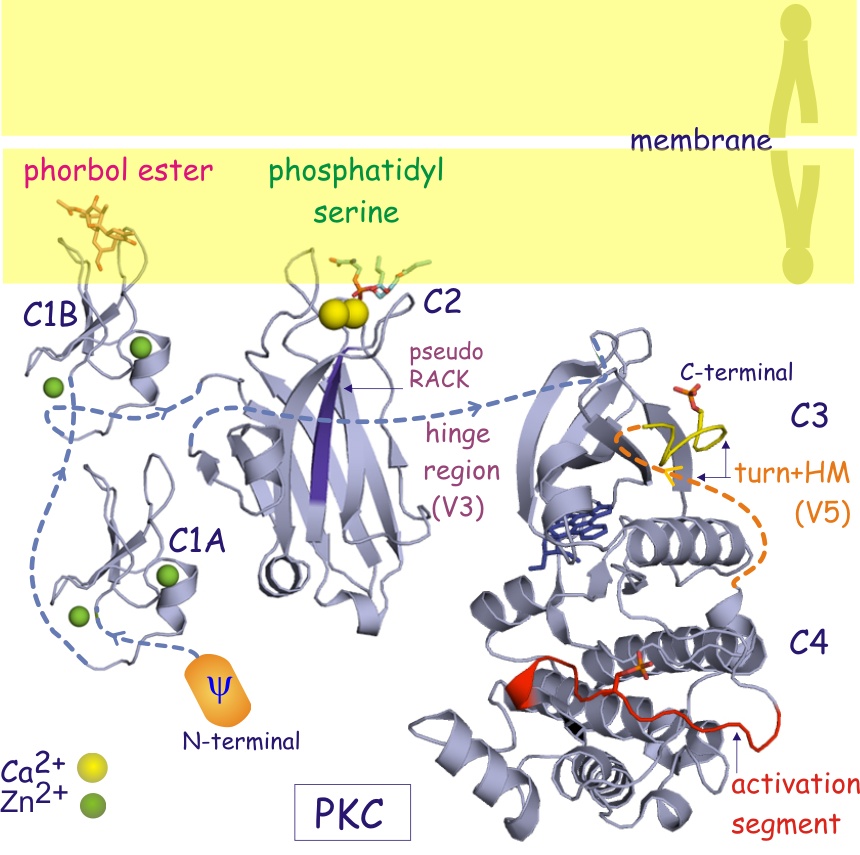
Componets of Protein kinase C consist of a single polypeptide with a N-terminal regulatory region and C-terminal catalytic region (Newton, 1995). These regions, containing approximately 20-40kDa and 45kDa respectively, are seperated by a hinge region in the core of the protein's sturcture. As previously stated, PKCs' subgroups differ by conserved regions in the protein as seen in Figure 2 below. Of these subgroups, conventional protein kinase Cs: α, βI, βII, and γ were the first to be discovered. This particular class differentiates itself because of its functional C2 region which is regulated by Ca2+ via a binding site formed by five aspartate residues in the core of the domain. Next researchers tackled novel protein kinase Cs: δ, ε, η (L), θ, and μ. These isozomes preserve the same structure as cPKCs, but the C2 domain no longer serves the same binding function as did cPKCs. Instead nPKCs rely more heavily on DAG for their full activiation. Finally are atypical PKCs: ζ and λ. This class, unlike the other two, varies quite differently in sturcture. Atypical PKCs' C1 domain only has one Cys-rich motif as well as their C2 domain no long contains any residues involved in maintaining the conformational fold found in both cPKCs and nPKCs (Newton, 1995).
Intrestingly, activation of PKC is complemented with the removal of pseudosubstrate (PS) from its kinase core region. This was shown to be true by incubating PKC with an antibody encoded for pseudosubstrate and thusly resulting in activation of the enzyme due to the removal of PS from PKC's active site (Newton, 1995). Additionally, ligands DAG and phorbol esters facilitate a change in the enzyme's membrane affinity that alters the surface hydrophobicity effectively causing membrane interaction despite confromational changes that may not occur between cPKCs and nPKCs (Newton, 1995).
Function:
Protein kinase C serves many vital roles in the cell and overall system. However, it mainly serves as a specific kinase enzyme for Ca2+, as designated by the C in PKC. While previously it was established that not all types of PKC use Ca2+ as a substrate for activation, it is known that PKC plays a pivitol role as a relay system in many Ca2+ dependent processes. Addittionally, PKC serves as a promoter of lipid hydrolysis as well as an effective key in signal transductions throughout the cell (Nishizuka, 1986). Generally, PKC's function is mediated by two mechanisms - 1) phosphorylations that align PKC for catalysis and localize it to the cytosol and 2) binding of substrates or ligands that free the PS from the core binding site thusly activating the enzyme.
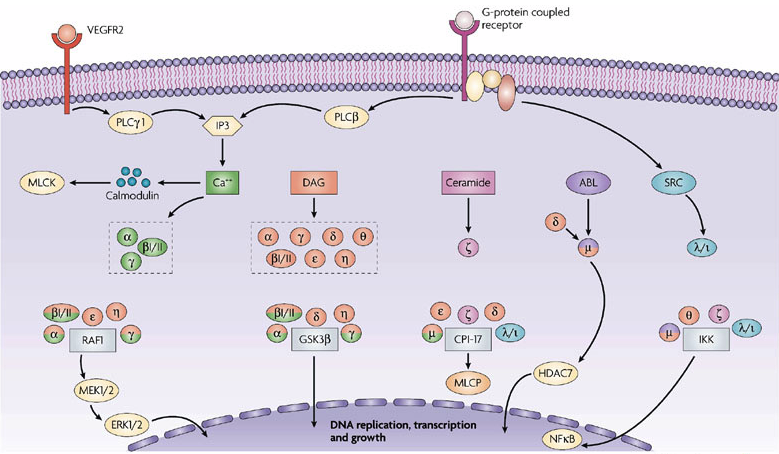
As for the function of PKC throughout the cell, many specualtions have evloved based off of various fields of research targeting PKC and its effect on different cells. In a study by Richard Whelan and Peter Parker on the effect of downregulation of cPKC, they found that the inhibition of PKC function resulted in induced apoptotic response (1998). Other researchers like Carsten Schmitz-Peiffer and Trevor J. Biden, found that insulin acted as a stimulant to cPKC and nPKC isoforms in the promotion of glucose disposal, a pivitol study that suggested a link between PKC and type 2 diabetes (2008). However because of its widespread capabilites presented through variously structured isozomes, PKC's functional expression is not necessarily clear (Steinberg, 2008). Nevertheless, it is sufficient to conclude that because of PKC's various structural differences due to locatoin of conserved domains that numerous enzymatic functions result from PKC interaction.
Works Cited
Newton A. 1995. Protein Kinas C: Structure, Function, and Regulation*. Journal of Biological Chemistry 270: 28495-28498.
Nishizuka Y. 1986. Studies and Prospectives on Protein Kinase C. Science 233: 305-312.
Schmitz-Peiffer C and Biden T. 2008. Protein Kinase C Function in Muscle, Liver, and β-cells and Its Therapeutic Implications for Type 2 Diabetes. Diabetes 57: 1774-1783.
Steinberg S. 2008. Structural Basis of Protein Kinase C Isoform Function. Physiological Reviews 88: 1341.
Whelan R and Parker P. 1998. Loss of Protein Kinase C Function Induces An Apoptotic Response. Oncogene 16: 1939-1944.