*This website was produced as an assignment for an undergratuate course at Davidson College.*
Hepatitis C Virus Non-structural Protein 3 helicase (NS3h)

This image shows the non-structural proteins of Dengue virus, which is thought to have a similar helicase to HCV. Image obtained from WikiCommons.
Hepatitis C Virus (HCV) General Information
Hepatitis C Virus is an enveloped, positive sense, single stranded RNA (ssRNA) virus that is a major cause of hepatocellular carcinoma, liver cirrhosis, and chronic hepatitis. There is no protective vaccine against HCV, and standard treatments are often ineffective or toxic to the patient. The HCV genome is 9.6kb in length and encodes three structural proteins (C, E1 and E2) and six non-structural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (1). Research into the structure and function of all nine proteins encoded by HCV is important to the development of potential novel drug therapies.
In order to replicate, HCV must first penetrate a host cell, which is generally a hepatocyte. Since HCV is an enveloped virus, a double fusion event allows the virus to gain entry to the cytoplasm of the host cell. After entering the host, the positive sense ssRNA is translated into proteins just as mRNA would be translated by the cell normally. One of these translated proteins is an RNA-dependent RNA polymerase that the virus uses to replicate its entire genome in order to create progeny viruses. The newly formed ssRNA genomes and proteins are combined to form new viruses that are released into the body by exocytosis allowing each new virus to obtain a cellular envelope (2). This description of the replication of HCV is very simplified, but it contains enough information to understand potential roles of non-structural 3 protein helicase in the HCV life cycle.
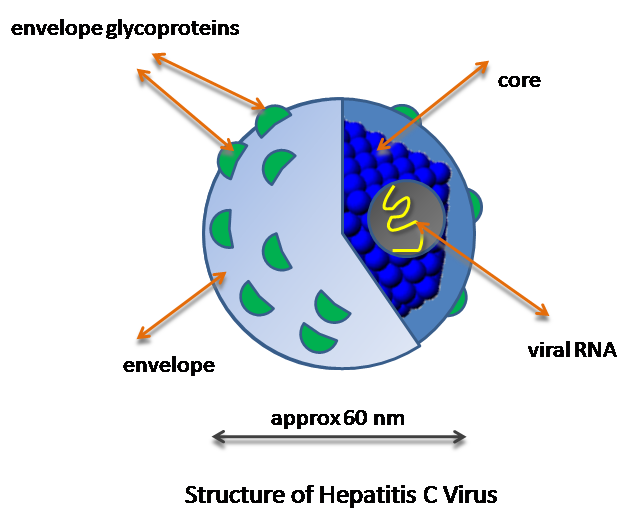
Image from WikiCommons
What is a helicase?
When a reference to the enzyme ‘helicase’ is made, generally, one is referring to DNA helicase. DNA helicase uses energy obtained from ATP hydrolysis to unwind DNA into two separate strands. The enzyme accomplishes this unwinding by moving or translocating along one strand of DNA in one direction. The movement of this enzyme interrupts the hydrogen bonding that holds the two strands of DNA together (3). Different helicases are capable of moving along either DNA or RNA. The strands of nucleic acids may be either double or single stranded. Additionally, the helicase may move in either a 3’ to 5’ or 5’ to 3’ direction. The basic function of any helicase enzyme is simply to translocate unidirectionally along a strand of nucleic acid. Because of this, there is a great diversity of functions for this enzyme including unwinding DNA, removing nucleic acid associated proteins, threading nucleic acids through openings during mRNA export, and participating in viral packaging/unpackaging. Because of the diverse capabilities of this protein, helicases are involved in almost all cellular processes involving nucleic acids (4).
What is NS3h?
NS3h stands for non-structural protein 3 helicase. NS3h is able to unwind both DNA and RNA in a 3’-5’ direction (5). The non-structural 3 protein of HCV is actually made up of multiple domains comprising two major sections. The N-terminal portion of the protein represents a serine protease that we will not discuss here. The C-terminal portion of the NS3 protein encodes a three-domain helicase. Because the life cycle of HCV does not include any steps involving DNA production, it may seem somewhat surprising that this virus encodes a helicase. Previous investigators have speculated that perhaps the helicase is involved in unwinding any RNA duplexes that form during the replication of the HCV genome. Additionally, the helicase could be used to flatten the structure of single stranded RNA to avoid any interruption to RNA-dependent RNA polymerase activity that could be negatively affected by the secondary structure of RNA (6). Regardless of the exact use of this helicase in replication of HCV, studies have shown that this protein is required for optimal replication of HCV (1).
.
Images A-F depict the HCV helicase structure from different views highlighting different domains and binding capabilities. A, C, E, and F highlight the secondary structure of the HCV helicase while B and D are electrostatic surface images which show the three dimensional shape of the surface of this protein. This figure is from an online book that is part of the National Library of Medicine public domain (6).
Structure/Function Information for NS3h
NS3h is made up of three domains. The most N-terminal domain is domain 1 while the C-terminal domain is 3 with domain 2 lying between the two. Flexible linkers connect domains 1 and 3 to domain 2 allowing domain 2 to rotate relatively freely. Various parts of the HCV helicase are conserved among many species while some portions are unique to only HCV and very closely related species. Additionally, even within the HCV genome, there is variation. For example, three different strains of the virus show a variation at residue 450. Strains 1 and 2 have a Thr at this position while 2a contains an Ile. When one of strains 1 or 2 have the Thr at position 450 replaced with an Ile, the protein is able to unwind DNA more quickly. It appears that this change from a Thr to an Ile is an adaptive mutation that allows the virus to perform more efficiently in a chimpanzee. This is because this mutation has only been seen in strain 2a, which was isolated from a chimpanzee while all human strains studied have a Thr at position 450. The portions of this protein that remain conserved among a wide range of species are related to a conserved function. For example, the portion of the protein encoding the ATP binding site is conserved among many species (6).
To view an animation that shows the movement of domain 2, click here.
Most research to date has focused on areas of HCV that are unique to this and related viruses with the hope that these domains can become targets for therapeutic studies. One of these regions is an Arg at position 393. This residue is hypothesized to be in contact with the nucleic acid that the helicase acts on. When the amino acid at this position is changed to an Ala, the helicase no longer unwinds nucleic acid strands. Another area of interest is a loop that extends from domain 2. In a highly conserved region of this loop, there is a pair of Phe at positions 438 and 444 that are of interest. Mutagenesis studies have shown that these Phe amino acids are important to the release of nucleic acids after ATP has bound to the enzyme. In addition to these sites of interest, all helicases that have been crystallized have domains that are similar to domains 1 and 2 in HCV. Nevertheless, no other helicase has shown similarity to domain 3 of HCV. When this domain was removed from NS3h, however, it was shown that domain 3 is essential for proper functioning of HCV (6).
The mechanism of action of NS3h has long eluded the scientists that study this protein. In a recently published study by Gu and Rice in 2009, crystallography was utilized to determine the translocation mechanism for this protein. In this study, the investigators were able to use a complex of ADP and BeF3 as well as ADP and AlF4- that mimic ATP hydrolysis. By visualizing the crystal structure of NS3h in simulated interactions mimicking ATP binding and hydrolysis, the researchers were able to form a hypothesis for helicase translocation. Investigators captured images of three distinct conformational states of this enzyme that suggest a rachet translocation mechanism (5). Investigators describe this rachet action as “the successive transitions of motif V and the spring helix release phosphate 3 from NABS1 and recapture it in NABS2, presumably preventing phosphate 3 from walking in the opposite direction” (7). This proposed mechanism would explain why this helicase is capable of unwinding DNA in the 3' to 5' direction, but not the reverse. Although it is difficult to imagine the exact mechanism of action for this protein without being an expert in structural biology, it is clear that nucleotide-dependent interactions between various domains of this protein are integral to the function of NS3h.
References
1. Mackintosh SG, Lu JZ, Jordan JB, Harrison MK, Sikora B, Sharma SD, Cameron CE, Raney KD, and Sakon J. Structural and Biological Identification of Residues on the Surface of NS3 Helicase Required for Optimal Replication of the Helpatitis C Virus. The Journal of Biological Chemistry 2006; 281 (6): 3528-3535.
2. Lindenback BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature 2005; 436: 933-938.
3. Purves WK, Sadava D, Orians GH, Heller HC. Life: The Science of Biology. 7 ed. New York, NY: Sinauer Associates and W. H. Freeman; 2003. 223 p.
4. Lüking A, Stahl U, Schmidt U. The protein family of RNA helicases. Critical Reviews in Biochemistry and Molecular Biology 1998; 33: 259-296
5. Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a rachet translocation mechanism. PNAS 2009; 107 (2): 521-528.
6. Tan, S, editor. Hepatitis C Virus: Genomes and Molecular Biology. Norfolk, UK: Horizon Bioscience; 2006. 207-244 p.
7. Gu and Rice 526.
Please direct questions or comments to my email kahasty@davidson.edu