*This website was produced as an assignment for an undergratuate course at Davidson College.*
Hepatitis C Virus Non-structural Protein 3 helicase (NS3h)
Orthologs discovered using BLASTp
Using the NCBI protein database, I was able to obtain the amino acid sequence for NS3h that was determined in a study by Gu and Rice in 2009 (1). Curiously, there was no nucleotide sequence for this protein. There were nine related entries in the NCBI nucleotide database, but all of these entries were very short (6-19 nucleotides) sequences that could not possibly account for the entire NS3h protein. The lack of a complete nucleotide sequence was not a concern because protein sequences are generally more highly conserved across species than nucleotide sequences anyway. Using the complete amino acid sequence, I performed a protein BLAST or BLASTp to compare the amino acid sequence of NS3h to a database of all amino acid sequences of all proteins for which the sequence is known. From this search, I obtained a short list of related helicase type proteins in other viruses. Below are images of some of the BLASTp results with short descriptions.

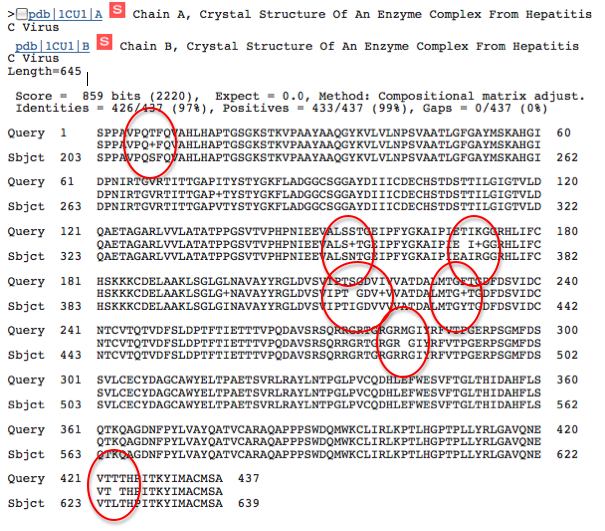
The first image is a very close match of the query sequence to 'an enzyme complex from Hepatitis C Virus.' Because of the very high conservation of this sequence and very low expect value, it is likely that the matching subject sequence is in fact the NS3h protein. It is interesting to note, however, that there are some differences in the sequence (circled in red). Perhaps the strain of Hepatitis C Virus used to determine the sequence for the subject is slightly different than the one used in the 2009 Gu and Rice study (1).
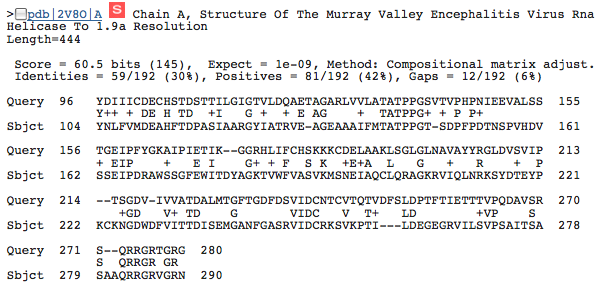
This image shows a portion of the query from amino acid 97 to 271 matching with a portion of the Murray Valley Encephalitis Virus RNA helicase. The expect value (1e-09) is low enough to suggest that this may be a conserved portion of the NS3h sequence that could be important to the helicase function of the NS3h protein.
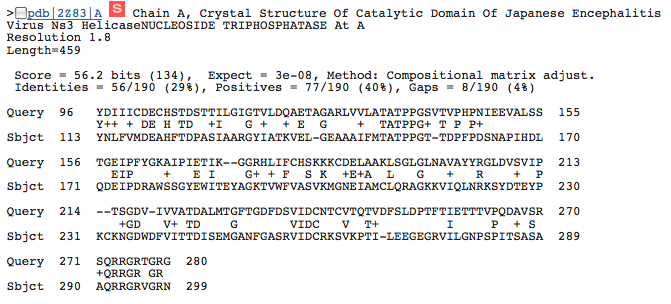
This image shows a match between a portion of the NS3h protein and a helicase for the Japanese Encephalitis Virus. Again, we find a low expect value (3e-08) that suggests this may be an important portion of the NS3h protein. Interestingly, the matching part of the query sequence is also from amino acid 96 to amino acid 271 just as it was for the match with the Murray Valley Encephalitis Virus.
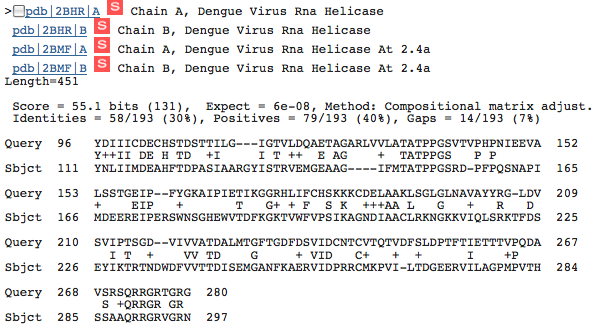
This image shows similarity between HCV NS3h and the Dengue Virus RNA Helicase. Again, we see the important amino acid 96-268 portion of the protein appears to be conserved.
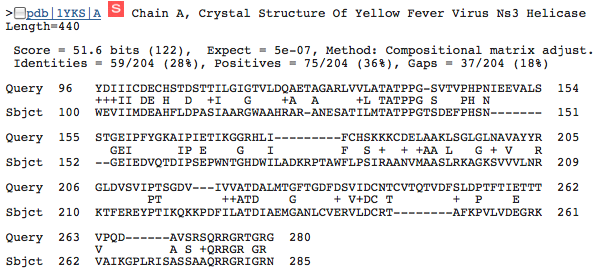
This image shows the similarity between HCV NS3h and a helicase from Yellow Fever Virus. Still we see that the 96-263 portion of the protein seems important to helicase function.
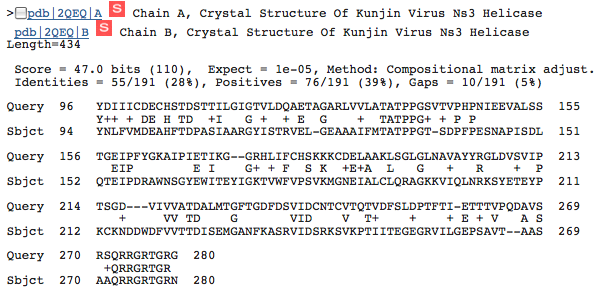
Finally, we see the same portion of the HCV NS3h protein match with a helicase from the Kunjin Virus.
NCBI conserved domain database
After reviewing the BLASTp data, it seems that the sequence between amino acid 96 and approximately amino acid 270 are very important to the helicase function of the HCV NS3h protein. In order to attempt to verify that the cause of this conservation of this portion of the sequence is in fact due to the helicase function of the protein, I searched the NCBI conserved domain database using amino acids 96-270 as a query sequence. There were two domain hits with significant e values that are shown below.
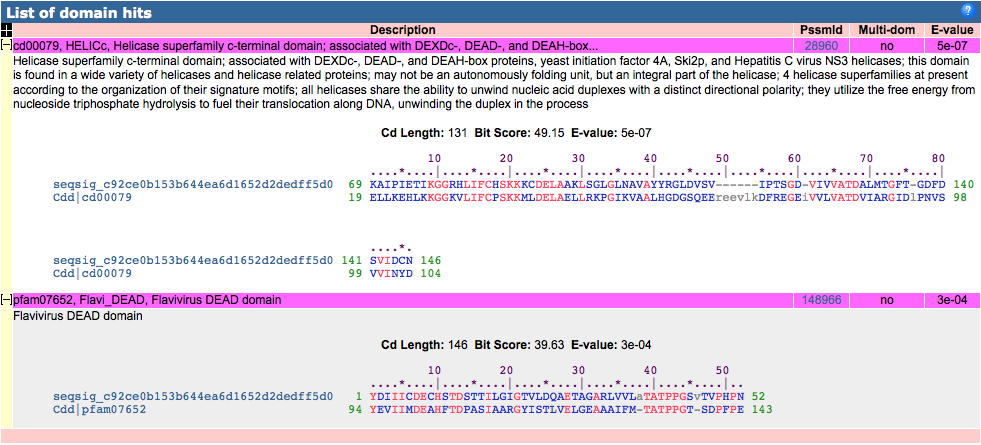
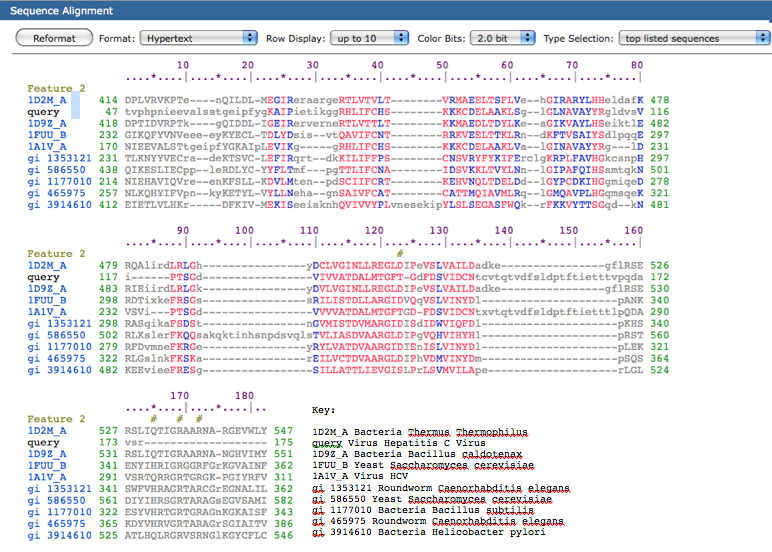
The top hit was the helicase superfamily C-terminal domain. This domain is associated with DEXDc-, DEAD-, and DEAH-box proteins, yeast initiation factor 4A, Ski2p, and of course the HCV NS3 helicases. NCBI describes this domain as an integral part of the helicase. Conserved features in this domain include a nucleotide binding site and an ATP-binding site, both of which are essential to the function of the helicase (unwinding DNA). Below is a sequence alignment generated by the conserved domain database. Exact residue alignments are shown in red, other alignments are in blue, and unaligned columns are in grey. In the bottom right of this figure is a key that has both the type of organism represented in each row and also the species represented. It is interesting to note that this conserved helicase domain is not only found in other viruses but also in bacteria, yeast, and even roundworms.
The second domain hit was for the DEAD-like helicase superfamily. This family includes a diverse array of helicases that use ATP to unwind either RNA or DNA. An ATP-binding site is part of this conserved domain. There is much less information available on the NCBI website for this superfamily than for the helicase superfamily C-terminal domain.
Viral evolution
Viral evolution is interesting to study because it not only involves the changes that accumulate in the viral genome over time but also involves several external factors that can influence the evolution of viruses. For example, if an anti-viral drug is used to combat a viral infection in a patient, the viral population in the individual patient may develop drug resistance due to a random mutation (or multiple mutations) that allows the virus to overcome the effects of the drug. If this patient then passes on the virus to a new host, the new viral infection will also be drug resistant. In addition to drug interactions, many viruses also can acquire new genes or nucleotide sequences due to insertion events in the host genome. If a virus undergoes a latancy period as part of its replication cycle, it may insert itself into the genome of the host organism and remain there until it is reactivated. When the viral genome pops back out of the host genome, it can rarely take host DNA with it which will then be incorporated into the viral genome. Because the helicase enzyme is so essential to basic DNA replication, different versions of this enzyme can be found in all known living organisms. It is therefore conceivable that some virus at some point in the evolutionary past picked up the DNA encoding this enzyme from a host organism. The original virus that acquired the helicase enzyme may not have been HCV. Because HCV does not have a latency period during its course of infection, no mechanism exists for HCV to pick up host DNA. However, the HCV helicase is closely related to four other viruses which may have a latency step in their replication cycle. Also, just as speciation occurs in other organisms, viruses can evolve from completely different viruses over long periods of time, so the helicase enzyme in HCV may have been incorporated into the HCV genome long before the virus formally became 'HCV'. Additionally, because of the similarity of important helicase domains among very varied species (yeast, bacteria, and roundworms), it is probable that this enzyme was found in a common ancestor to all these species at one point in time. Therefore, it is difficult to say from which host species the viral helicase enzyme orginally came from. Regardless of which virus acquired the helicase enzyme from which host, there is a mechanism by which the virus could acquire the helicase enzyme from a host. Because of the existance of a mechanism whereby the HCV helicase enzyme could have been obtained from a host organism as well as the data that show similarities between the HCV helicase amino acid sequence and sequences from varied species, it is likely that the HCV helicase enzyme was acquired from a host organism in the evolutionary past.
References
1. Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a rachet translocation mechanism. PNAS 2009; 107 (2): 521-528.
Please direct questions or comments to my email kahasty@davidson.edu