* This webpage was created as an undergraduate assignment at Davidson College, Davidson, NC*
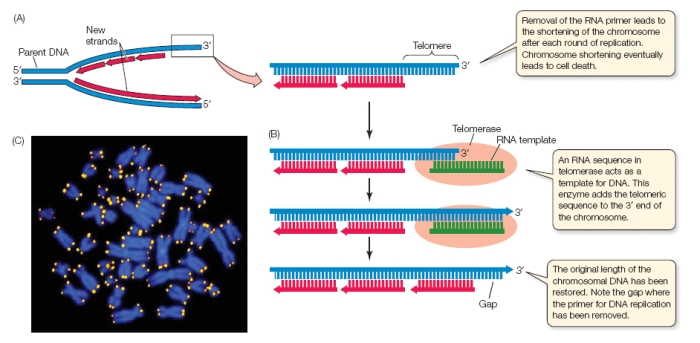
Eukaryotes have evolved ways to overcome restrictions in DNA replication mechanisms. Unlike prokaryotes, they do not have a single circular chromosome but rather multiple linear chromosomes. One of the difficulties facing eukaryotes is that they are unable to replicate the ends of their chromosomes, known as the telomeres, which results in the shortening of chromosomes over time and the removal of essential DNA information. The shortening of chromosomes occurs because the DNA polymerase can only add 5’ to 3’ on a given strand of the DNA during replication. Helicase unwinds the DNA to separate the two strands of single stranded DNA so that they can each act as templates. The leading strand is able to add nucleotides 5’ to 3’ for the length of the chromosome. The lagging strand must also add 5’ to 3’ but the strand is antiparallel and the DNA polymerase can only add to existing RNA primers. At the very end of the chromosome, there is a sequence of DNA that cannot be replicated because there is no RNA primer at the 3’ end (Figure 1). This cell division problem has been remedied in eukaryotes by the production of telomerase, which lengthens the telomeres by adding a repeated sequence to the end of the template.

Figure 1. The diagrammed mechanism of telomerase. A) A eukaryotic replication complex where the top strand is the lagging strandand and the bottom strand is the leading strand. The telomere not able to be replicated due to the lack of RNA primer, which results in the shortening of chromosomes with many cell divisions. B) Telomerase places an RNA template that links with the telomeric DNA. The enzyme will add the telomeric sequence to the 3' end in order to lengthen the chromosome. C) Eukaryotic chromosomes (blue) are fluorescently stained to show the location of the telomeres (yellow) using the conserved repeat sequence.
© 2008 by Sinauer Associates. Permission Pending.
For an animated step by step explanation of how telomerase works, see the Telomerase Animation provided by Rutgers University
Telomerase contains an RNA component that acts as a template for the telomeric repeat sequence. The telomere sequence in humans is TTAGGG and is repeated 2,500 times (Purves et al., 2004). Human chromosomes lose about 50 to 200 nucleotides with each cycle of DNA replication and cell division (Purves et al., 2004). Telomerase in germ cell lines acts to “reset” the telomeres and can also be found in rapidly dividing bone marrow (Purves et al., 2004). However, it becomes dormant in somatic cells as the individual enters adulthood. A normal individual’s telomeres shorten with age because of the cell division problem and the natural inactivity of telomerase (Gillis et al., 2008). A cell can only divide so many times before enters apoptosis or programmed cell death due to deleterious deletions in the coding regions of DNA. The presence of telomerase in somatic cells has been linked to over 90% of human cancers (Purves et al., 2004). Thus, it may confer a type of immortality to rapidly dividing carcinogenic cells.
Telomerase functions as both a dimer and as a monomer, but was isolated and characterized as a monomer. It is composed of two subunits, known as TER and TERT. TER is the RNA template subunit that contains the sequence 3’-AACCCCAAC-5’ (Romi et al., 2007). TER varies in size, sequence and structure among species but its core structural elements are conserved (Gillis et al., 2008). TERT is the highly conserved protein catalytic subunit and is closely related to viral reverse transcriptase (Gillis et al., 2008). TERT has three domains and forms a doughnut shape (Figure 2). One domain is the high-affinity TER binding domain (TRBD). TRBD contains mostly alpha-helices and an indentation on its surface created by two conserved motifs, CP and T (Gillis et al., 2008). CP is known to bind double stranded RNA and T binds single stranded RNA of the TER template. The reverse transcriptase domain has a mixture of alpha-helices and beta-strands and contains both the ‘fingers’ and ‘palm’ subdomains. Lastly, the CTE domain acts as the ‘thumb’ of TERT. It is composed of an elongated helical bundle and several long loops at the surfaec (Gillis et al., 2008).

Figure 2. The three domains of the catalytic subunit TERT of telomerase. The TRBD is the high-affinity TER binding domain and contains mostly alpha-helices (purple). The reverse transcriptase domain contains both the palm and fingers sub domains, composed of a mixture of alpha helices and beta strands (tan and orange). The thumb domain is the CTE domain that has an elongated helical bundle and long loops at the surface (red).
©2007 Gillis et al. Permission Pending.
For the full three dimensional structure of telomerase, see the PDB website of Telomerase by Gillis et. al
The space within the TERT ring is functionally important because it allows for the RNA-DNA interaction. In general, the organization of motifs resembles the spiral double helix of double stranded nucleic acids (Figure 3a). The motifs in contact with the DNA are positively charged and extend into the hole to interact with the negatively charged DNA backbone (Gillis et al., 2008).
The ‘thumb’ domain maintains the three dimensional structure of DNA. The thumb loop connecting the ‘thumb’ to ‘palm’ domain, known as the primer grip, corresponds to the double helix structure. Lysine and asparagine, both positively charged amino acid residues, are directed towards the center of the TERT molecule where they will interact with the DNA backbone. Specifically, the lysine at K406 was hypothesized to interact directly with the DNA model to facilitate the placement of the incoming 3' DNA strand (Figure 3b). Additionally, the alpha helix 19 is located at a minor groove of the RNA/DNA model, which may assist the RNA-DNA hybrid binding (Figure 3b). A deletion of this motif resulted in severe loss of TERT activity (Gillis et al., 2008).
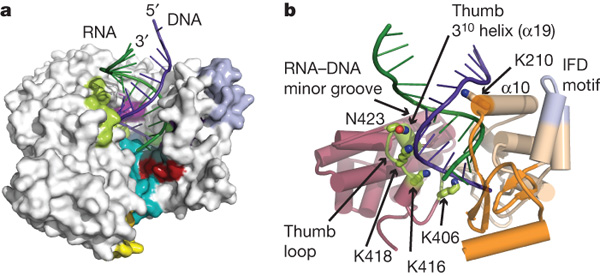
Figure 3. a) The interaction of TERT, the RNA template and the growing DNA strand. b) The thumb domain perfectly fits the double helix structure of DNA. K418, K406 and K418 are all lysine residues that stabilize the 3' negatively charged DNA backbone due to their positive charges. The alpha helix 19 interacts with the RNA-DNA minor groove to stabilize the RNA binding to DNA.
©2007 Gillis et al. Permission Pending.
The binding pocket of the active site is located between the ‘fingers’ and ‘palm’ domains. The active site contains three necessary aspartic acid residues. The surrounding motifs are speculated to be involved in binding the template to the nucleotides. The hydrophobic pocket adjacent to the three catalytic aspartates would accommodate the similarly hydrophobic nucleotide bases (not shown). This hydrophobic pocket would place the triphosphate of the nucleotide substrate near the catalytic site with Mg2+ (not shown). At the same time, the ribose of the nucleotide substrate is positioned at the RNA-binding pocket near a glutamine, which has been shown to increase enzyme specificity (Gillis et al., 2008).
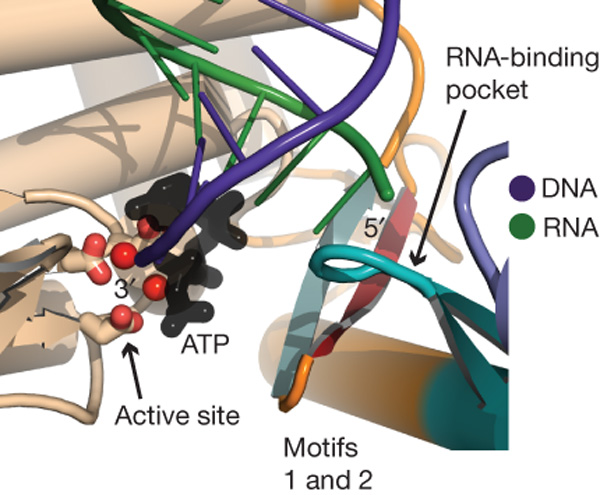
Figure 4. The motifs of the 'fingers' and 'palm' domains support the active site. The DNA strand (in purple) would be located at the active site given the position of the hydrophobic pocket near the active site that can accommodate the hydrophobic bases. The positioning of the DNA near the active site is essential in order for the enzyme to add nucleotides to the growing chain of DNA. The RNA strand (green) interacts with the RNA-binding pocket, which was found to contain glutamine, thus increasing enzyme specificity. The ß-hairpin (red and green) of the TRBD is closely associate with both the thumb loop and motifs 1 and 2.
©2007 Gillis et al. Permission Pending.
TRBD binds to the TER template and positions it near the active site. The domain forms a cavity on the side of the ring (Figure 2), which allows for the placement of the RNA template inside the ring at the active site. A ß-hairpin extends from the RNA-binding pocket and interacts with the thumb loop and motifs 1 and 2 (Figure 4). The researchers hypothesized that the ß-hairpin may act as an allosteric switch to couple RNA binding and the placement of the RNA template at the active site. Proper placement of the RNA template would facilitate pairing with the DNA substrate. Additionally, it may help to explain “repeat addition processivity,” which is the telomerase-specific feature that allows for multiple additions of telomeric sequences. (Gillis et al., 2008).
The function of telomerase has been intimately linked to its structure. The ring structure of telomerase closely resembles that of reverse transcriptase, suggesting that they are both evolutionarily and functionally associated. The RNA-DNA model complex in the interior of the TERT subunit places the 3' DNA at the catalytic site, where it is available for nucleotide polymerization. The RNA strand is expect to interact with the RNA-binding pocket, where the TER and TERT subunits meet. This model suggests a mechanism for the association of the RNA template and DNA substrate in the activity of telomerase.
©2010 Danielle Jordan, Biology Department, Davidson College, Davidson, NC
Questions? Comments? Email me at dajordan@davidson.edu