*This website was produced as an assignment for an undergratuate course at Davidson College.*
HUMAN SERUM ALBUMIN
What is Human Serum Albumin?
Human serum albumin (HSA) is the most abundant protein in blood plasma, accounting for nearly 60% of the total protein in blood serum and showing a typical blood concentration of 50g/L. Albumin, which is synthesized in the liver, is a soluble, monomeric, globular protein with a molecular mass of 67 kDa. HSA has an exceptional binding capacity for a wide variety of hydrophobic ligands and plays a key role in the transport of fatty acids, amino acids, metals such as Cu2+ and Zn2+, metabolites such as bilirubin, and pharmaceutical drug compounds. The most essential physiological role of the HSA carrier protein is therefore thought to be the transport of such water-insoluble solutes in the bloodstream to target organs such as the liver, intestine, kidney, and brain (Sugio et al., 1999). As a result, HSA contributes significantly to the regulation and maintenance of the extracellular fluid levels as well as the the pH and osmotic blood pressure of the plasma (He et al., 1992).
View the crystal structure of human serum albumin in Jmol (Human Serum Albumin). RCSB Protein Data Base.

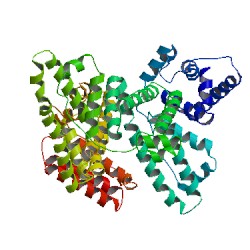
Image from RCSB Protein Data Bank. Permission pending.
Figure 1: Crystal Structure of human serum albumin showing the helical structures and the six subdomains, each of a different color.
Structure and Function of HSA
Domains (I, II, III): The primary sequence of HSA shows that the protein is a single polypeptide of 585 amino acids residues containing 17 pairs of disulfide bridges, one free thiol (Cys 34), and a single tryptophan (Trp 214). The unglycosylated protein is composed of three structurally similar, though asymmetrical, repeating domains—I (residues 1-195), II (196-383), and III (384-585)—each of which are divided into two subdomains, A and B. This series of six helical subdomains assembles to form a heart-shaped molecule (Figure 2; Sugio et al., 1999). Domains I and II are almost perpendicular to each other in which the tail of subdomain IIA is attached to the interface region between subdomains IA and IB by hydrophobic interactions and hydrogen bonds. Conversely, domain III projects out from subdomain IIB at a 45º angle to form a Y-shaped configuration for domains II and III. Thus, domain III only interacts with subdomain IIB (He et al, 2002). The HSA protein has been found to be very flexible in different conditions and can easily change its shape when participating in binding interactions due to the relative motions of the domain structures. The relative arrangements of the three domains are largely changed when fatty acid chains bind to the protein. During complex formation, the domains largely retain their conformation; however, the two C-terminal helices in domain III move toward the outside of the molecule (Sugio et al, 1999).
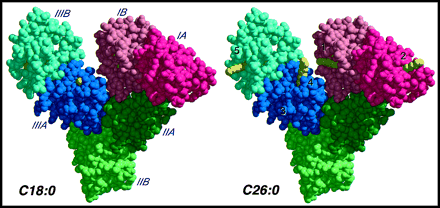
Image from Choi et al, 2002. Permission pending.
Figure 2: X-ray crystal structure showing the three domains (I, II, and III) and two subdomains (A and B) of HSA. On the left with C18:0 (carbon chain of 18C), fatty acids bind within the hydrophobic core of the subdomain binding sites on HSA and are not visible in this model. On the other hand, the methylene tail (yellow) of C26:0 protrudes largely into the water in four of the binding sites, suggesting that HSA binding sites are not suitable for long-chain fatty acids of C26.
Subdomains (A and B): In each domain, there are 10 principal helices (Figure 1; Figure 4). Subdomain A consists of a cluster of four α-helices (A-h1 to A-h4) flanked by two short α-helices (A-h5 to A-h6) tied together by a pair of disulfide bridges, while subdomain B has an additional cluster of four α-helices (B-h1 to B-h4). A long extended loop crosses the two subdomains to link them together within each domain. In total, there are 28 helices rather than 30 in the HSA molecule, due to the fusion of the last helices (B-h4) of domains I and II with the first helices (A-h1) of the next domains. Within HSA, 35 cysteine residues exist, 34 of which form 17 disulfide bridges, folding and looping together the α-helices into a unique 3-D structure. The disulphides are positioned in a repeating series of nine loop-link-loop structures which are centered around eight sequential Cys-Cys pairs. The N-terminal portions of the B subdomains complement the helical patterns by extending the polypeptide, forming what appears to be a simple up-down helical bundle (Sugio et al., 1999). Except for the IA-h3 subdomain, each subdomain forms a hydrophobic core. Amino acids having aliphatic and aromatic side chains cluster at the inner surface of the helix wall, forming deep crevices in the protein where fatty acids and other water-insoluble molecules can bury their carbon-rich tails safely away from the surrounding water environment (Choi et al., 2002).
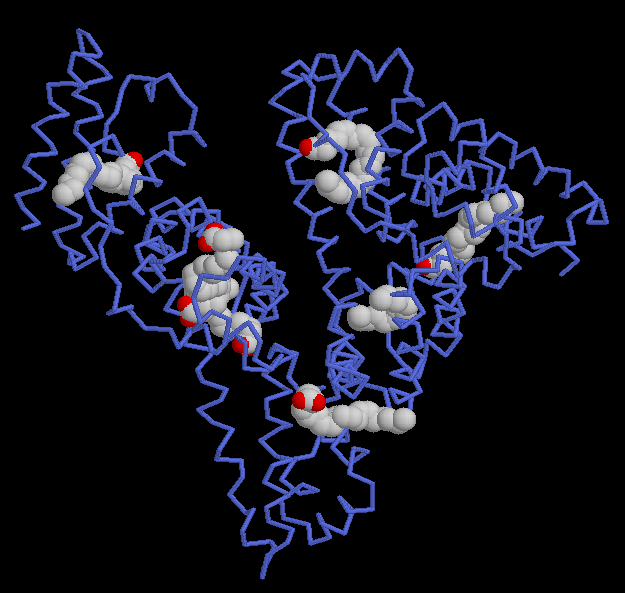
Image from RCSB Protein Data Bank. Permission Pending.
Figure 3: Seven molecules of arachidonic acid bind to the HSA protein. The protein is shown with blue tubes and the fatty acids are shown with spheres at each atom. The protein chain surrounds the carbon-rich tails of the fatty acids, shielding them from the surrounding water. Arachidonic acid is an unsaturated fatty acid with several double bonds that form rigid kinks in the carbon chain. It is important for the construction of molecular messengers used to signal pain and inflammation, and HSA can transport this fatty acid to its target destination through the plasma.
Ligand Binding: It is the exceptional propensity of HSA to bind to and carry fatty acids, steroids, metals, and other water-insoluble molecules that makes its structure so important. Generally, HSA has a greater affinity for small, negatively charged hydrophobic molecules (He et al., 1992). Each HSA molecule can carry up to seven fatty acid molecules (Figure 3; Goodsell, 2003). Evidence has shown that HSA contains six dominant areas of ligand association. There are two high affinity binding sites for small heterocyclic or aromatic compounds (located on subdomains IIA and IIIA), two to three dominant long-chain fatty acid binding sites (located on subdomains IB and IIIB), and two distinct metal-binding sites, one involving Cys-34 and the other the N terminus (Figure 4; Sugio et al., 1999 and Curry et al., 1992).
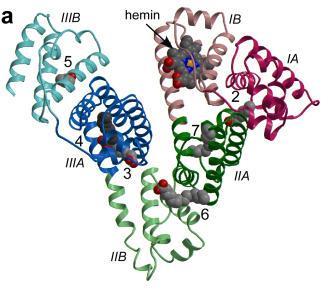
Image from Zunszain et al, 2003. Permission pending.
Figure 4: The secondary structure of HSA is shown with the subdomains color-coded. The location of ligand binding on HSA is indicated. Ligands are shown in a space-filling representation, coloured by atom type: carbon--gray; nitrogen--blue; oxygen--red; iron--orange. Fatty acid binding sites are labeled 2-7.
Since many ligands have been found to bind preferentially to IIIA, the binding cavity of this subdomain is the most active and accommodating on HSA (Figure 5; Dockal et al., 1999). Subdomains IIA and IIIA are the locations for the primary fatty-acid and bilirubin-binding sites. At similar positions in their structures, subdomains IIA and IIIA contain deep pockets lined with hydrophobic side chains surrounded by the first few residues of an extended loop together with helices A-h5 and A-h6. The entrance to the pocket in both IIA and IIIA is surrounded by positively charged amino acid residues (Sugio et al., 1999). Strategically located in the hydrophobic pockets of IIA and IIIA, residues Trp 214, Lys 199, and Tyr 411 are suspected by several studies to be crucial to the binding process by limiting the solvent accessibility (Sugio et al., 1999 and He et al., 1992). The crystal structure of HSA shows that domains II and III share a common interface, explaining why ligands bound to domain III affect conformational changes and binding affinities in domain II (Figure 2). Unlike IIA and IIIA, subdomain IA does not have a pocket near its hydrophobic core due to an extended polypeptide after Cys 62, and thus, because of its structural differences, IA is not an active ligand binding site (Sugio et al., 1999).
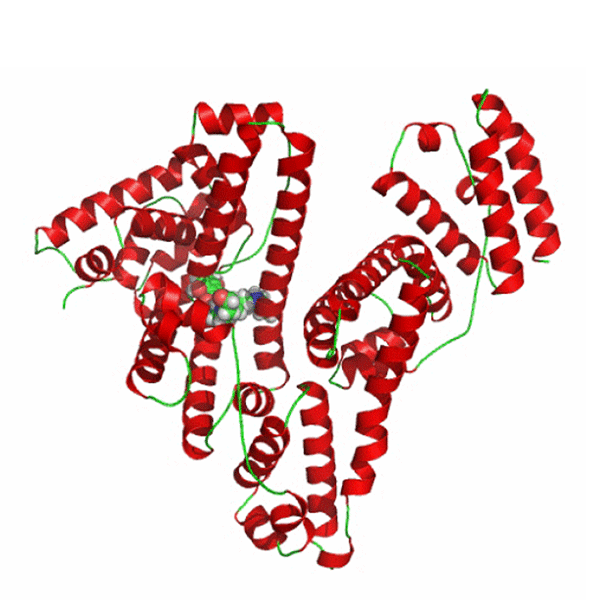
Image from Bhattacharya et al., 2000. Permission pending.
Figure 5: Mitoxantrone (MTX) is a clinically used antitumor anthracycline, which is transported to the target tissues by HSA. MTX is bound to the IIIA subdomain, the binding cavity with the highest affinity for ligands on HSA. The binding pocket is lined by hydrophobic side chains and the double disulfide bridges of helix IIIA-h3.
References
Bhattacharya AA, Grune T, Curry S. Crystallographic analysis reveals common modes of binding of medium and long-chain fatty acids to human serum albumin. J Molecular Biology 2000; 303: 721.
Choi JK, Ho J, Curry S, Qin D, Bittman R, Hamilton JA. Interactions of very long-chain saturated fatty acids with serum albumin. Journal of Lipid Research 2002; 43:1000-1010.
Curry S, Mandelkow H, Brick P, Franks N. Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nature Structural Biology 1998; 5: 827-835.
Dockal M, Carter DC, Ruker F. The three recombinant domains of human serum albumin: structural characterization and ligand binding properties. The Journal of Biological Chemistry 1999; 274: 29303-29310.
Goodsell DS. Serum albumin. January Molecule of the Month. RCSB Protein Data Bank 2003.
He XM, Carter DC. Atomic structure and chemistry of human serum albumin. Nature 1992; 358: 209-215.
Sugio S, Kashima A, Mochizuki S, Noda M, Kobayashi K. Crystal structure of human serum albumin at 2.5 Å resolution. Protein Engineering 1999; 12: 439-446.
Zunszain PA, Ghuman J, Komatsu T, Tsuchida E, Curry S. Crystal structural analysis of human serum albumin complexed with hemin and fatty acid. BMC Structural Biology 2003; 3:6.
Please direct questions or comments to Sarah Little