Structure and Function
Beta-1,4-Endoglucanase is a specific enzyme that catalyzes the hydrolysis of cellulose. It is produced chiefly by fungi, bacteria, and protozoans.
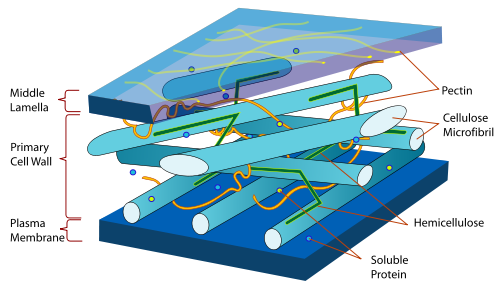
Cellulose is one of the main components of the plant cell wall. The xylem tissue mainly consists of cellulose which is furthur protected by hemicellulose and pectin (Figure 1) (http://en.wikipedia.org/wiki/Cell_wall , 2007).
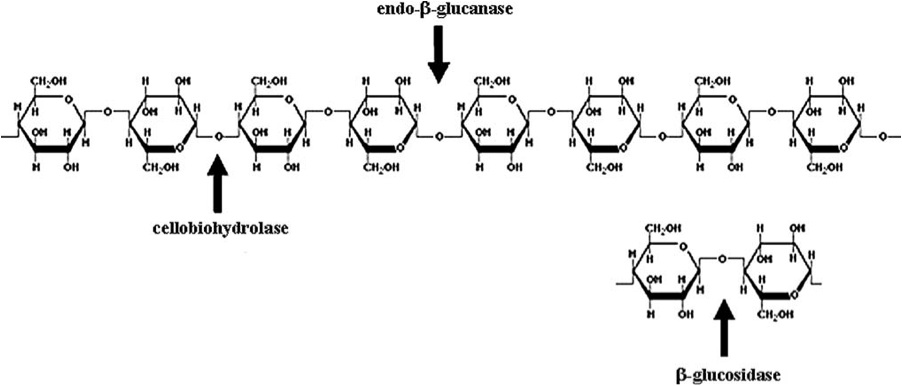
Beta-1,4-glycosidic bonds (hence the name, beta-1,4-endoglucanase) link together the beta-D-glucopyranose units of cellulose. Beta-1-4-endoglucanase enzymes specifically cleave the internal bonds of the cellulose chain. Exoglucanases and beta-glucosidases are needed to completely break down cellulose into glucose monomers (Figure 2) (Kumar, et al., 2008).
The Endoglucanase Complex
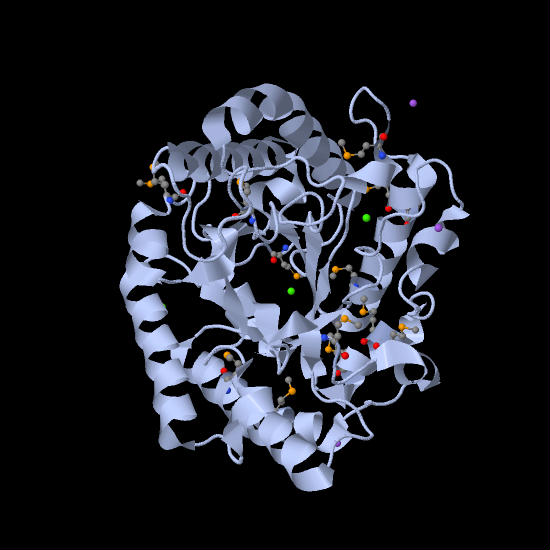
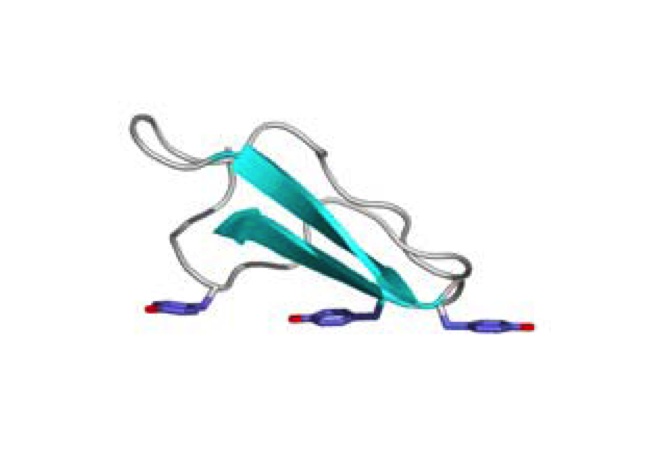
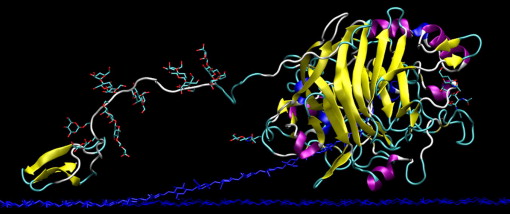
In the past 20 years, researchers have found that endoglucanases cannot break down polysaccharides efficiently without the help of non-catalytic carbohydrate-binding modules. Thus, endoglucanase is mostly found in the form of a complex that is made up of three separate domains. The main domain contains the large, globular catalytic domain which expresses the active site. A loop of the protein chain forms a tunnel that encloses the active site. This is attached at the O-glycosylated B block hinge region of the catalytic domain to the smaller, globular CBM at its C-terminal A block by a linker peptide made up of proline, serine, and threonine (Nimlos, et al., 2007).The overall shape of the complex looks like a tadpole, with the A and B blocks forming the extended tail and the catalytic domain forming the head (Pilz, et al., 1990). The CBM facilitates the enzyme by binding the complex to the cellulose, thus maintaining the proximity of the enzyme and the substrate. It can also target areas of the cellulose that are specific to the enzyme complex. In addition, the CBM itself can disrupt the structure of the cellulose and thus expose the substrate more to the enzyme. The aromatic amino acid residues and planar architecture of the CBM binding sites are complementary to the hydrophobic sites of cellulose chains (hydrophobic 110 face) (Figure 3). This exposes the beta-D-glucopyranose rings in the chair conformation, which have their alpha and beta faces having either two or three axial hydrogens exposed and ring hydroxyl groups in the equatorial position, allowing the CBMs to bind efficiently (Nimlos, et al., 2007). The thermodynamic forces that drive this interaction is controversial, but most researchers postulate that it comes from the positive entropy when the water molecules are released from the protein and ligand (Boraston, et al., 2004).
Mechanism
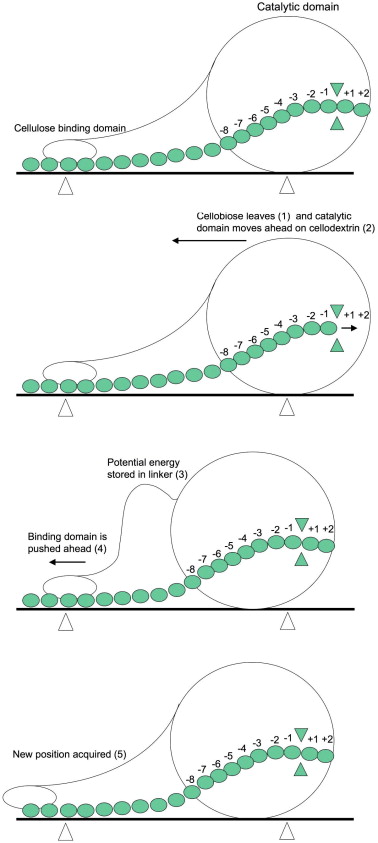
The endoglucanase complex hydrolyzes the cellulose chain in a processive manner from the reducing end of the chain (Figure 4) (Zhong, et al., 2009).
The CBM "pulls up" the chain and feeds it into the catalytic domain (Figure 5) (Zhao, et al., 2008). As aforementioned, the CBM's hydrophoic binding site of three tyrosines remains in contact with the surface throughout the reaction but can freely move translationally. The CBM undergoes a structural conformation after the substrate is in place. The fourth tyrosine (Y13) unfolds from within the CBM and forms a van der Waals interaction with the cellulose surface on the other side of the chain, thus encompassing the reducing end. While the rest of the CBM structure remains fairly rigid because of hydrogen bonds (between strand beta-3 and beta-1 and beta-2) and disulfide bridges (between beta-1 and beta-2) that maintain spacing matching that of the cellulose monomers at the binding site, the fourth tyrosine is located on the remainder of a loop of the protein and is more flexible for the induced fit (Nimlos, et al., 2007). Within the active site, Glutamine (212) acts as a catalytic nucleophile while Trypothan (40) fixes the substrate by hydrophoic interaction at the entrance of the active site tunnel. Similar to myosin and other motor proteins, the enzyme complex is driven by the hydrolysis of the glycosidic bond of cellulose (like phosphate bonds in ATP) as a source of energy for movement along their substrate (Igarashi, et al. 2009).
Relevance
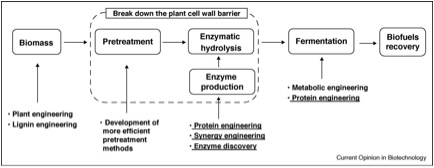
The increasing dependence of modern society on fossil fuels has resulted in an intensive search for alternative sources of fuel. The issue is of grave concern because of the environmental, social, and economic implications inherent in continued consumption of unsustainable fuels and sources of energy. Global climate change will continue to occur due to the increasing effect of carbon emissions in the atmosphere, political clashes will continue to grow over the control and allocation of these depleting resources, and the costs of these fuels will undoubtedly continue to rise (Merino S.T., and Cherry, J., 2007) Lignocellulosic biomass (second-generation) carbon resources accumulated from agriculture, forestry, and other industries are a promising, renewable, and globally accessible alternative to petroleum, which liquid transportation fuels currently are almost entirely derived from (Gibbons, W.R., and Hughes, S.R., 2009). The bioconversion process uses enzymes such as endoglucanase to break down cellulose into sugars that can be fermented into ethanol (Figure 6) (Wen, et al., 2009). Researchers have been looking to engineer microorganisms to withstand the harsh conditions of the process and to produce the necessary enzymes abundantly. It is necessary to understand these enzymes more fully and to improve them via protein engineering so that biomass can be efficiently and inexpensively converted into biofuels.
References
Boraston, A.B., Bolam, D.N., Gilbert, H.J., Davies, G.J. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769-781.
Gibbons, W.R., Hughes, S.R. 2009. Integrated biorefineries with engineered microbes and high-value co-products for profitable biofuels production. In Vitro Cell Dev Biol. 45: 218-228.
Igarashi, K., Koivula, A., Wada, M. Kimura, S., Penttila, M., Samejima, M. 2009. High speed atomic force microscopy visualizes processive movement of Tricoderma reesei cellobiohydrolase I on crystalline cellulose. Journal of Biological Chemistry. 284(52): 36186–36190.
Kumar, R., Sing, S., Singh, O.V. 2008. Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives. J Ind Microbiol Biotechnol. 35: 377-391.
Merino, S. Cherry, J. 2007. Progress and challenges in enzyme development for biomass utilization. Adv Biochem Engin/Biotechnol. 108: 95-120.
Nimlos, M.R., Matthews, J.F., Crowley, M.F., Walker, R.C., Chukkapalli, G., Brady, J.W., Adney, W.S., Cleary, J.M., Zhong, L., Himmel, M.E. 2007. Molecular modeling suggests indcued fit of Family 1 carbohydrate-binding modules with a broken-chain cellulose. Protein engineering Design and Selection. 20(4):179-187.
Pilz, I., Schwarz, E. Kilburn, D.G., Miller, R.C. Jr., Warren, R.A.J, Gilkes, N.R. 1990. The tertiary structure of a bacterial cellulase determined by angle X-ray-scattering analysis. Biochem. J. 271:277-280.
Wen, F., Nair, N.U., Zhao, H. 2009. Protein engineering in designing tailored enzymes and microorganisms for biofuels production. Current Opinion in Biotechnology. 20: 412-419.
Zhao, X., Rignall, T.R., McCabe, C., Adney, W.S., Himmel, M.E. 2008. Molecular simulation evidence for processive motion of Tricoderma reesei Cel7A during cellulose depolymerization. Chemical Physics Letters. 460: 284-288.
Zhong, L., Matthews, J.F., Hansen, P.I., Crowley, M.F., Cleary, J.M., Walker, R.C., Nimlos, M.R., Brooks III, C.L., Adney, W.S., Himmel, M.E., Brady, J.W. 2009. Computational simulations of the Trichoderma reesei cellobiohydrolase I acting on microcrystalline cellulose 1beta: the enzyme-substrate complex. Carbohydrate Research. 344(15):1984-1992
Web References
Wikipedia Plant Cell Wall Entry
Links
Please direct questions or comments to brpearson@davidson.edu