What the Literature Says about the Orhtologs of Calmodulin
Eukaryotes:
Basic databases such as Entrez Gene and GeneCards list various and diverse organisms in which the calmodulin gene is conserved, including: chimpanzees, dogs, cows, mice, fruit flies, worms, yeast, frogs, chicken, zebrafish, rice and thale cress (a small flowering plant). Research papers surrounding various facets of the calmodulin gene and protein tell a much deeper story.
In fact, Stevens notes that calmodulin has been found in ALL eukaryotic cells, which have been examined, noting cows, sea anemones and scallops specifically. Other sources assert that calmodulin has been found in ALL mammals, birds and amphibians (Nojima, 1987). Lagace et al. claim that calmodulin is present in all eukaryotic cells, including plants and protozoan (Tetrahymena pyriformis) (Lagace et al., 1982). Overall, the literature supports the “ubiquitous distribution” of calmodulin in eukaryotes, therefore indicating that orthologs of calmodulin exist in most eukaryotes (Baba et al., 1984). Table 1 supports the extreme distribution of the calmodulin protein within various species.
The literature not only agrees on the ubiquitous existence of the calmodulin protein in various eukaryotes, but the literature also cites the extremely high conservation of calmodulin protein sequence in these diverse organisms. For instance, Stevens highlights that calmodulin is highly conserved biologically and chemically (Stevens, 1982). In fact, Stevens notes that when comparing the calmodulin sequence from a cow and a sea anemone, only 3 amino acid residues differ between these three extremely different eukaryotes. This little difference suggests high conservation of the calmodulin protein.
Lagace et al. also explores conservation of the calmodulin protein sequence. They note that within the all mammalian species the calmodulin protein sequence is identical, whereas invertebrate calmodulin sequences differ from mammalian in only 3 amino acid residues (as previously shown by Stevens) and the protozoan calmodulin sequence differs in only 11 amino acid residues from the mammalian. It is critical to realize that the calmodulin sequence is 148 amino acids long, therefore these differences are slight and support high protein sequence conservation across distinct species. Klee et al. also support the strong conservation of the primary amino acid sequence of calmodulin by indicating that at most invertebrate calmodulin protein sequences differ from the vertebrate calmodulin sequences between 5-11 amino acids, which are usually at the carboxyl terminus of the protein.
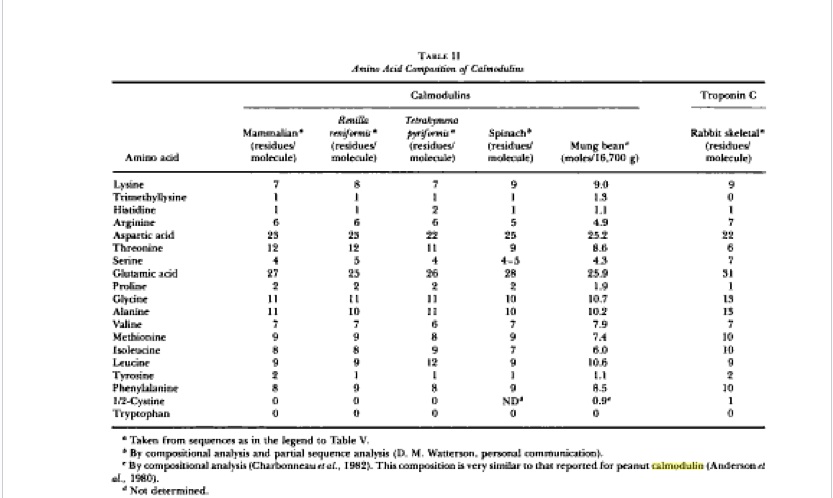
Not only does the literature explore sequence conservation, but Klee et al. also explore amino acid residue conservation in terms of similar numbers of different amino acids in each molecule of calmodulin. Table II denotes the number of different amino acids within 1 molecule of calmodulin.
The table above shows that the number of each type of amino acid within the calmodulin sequence of different organisms is conserved strongly. Most of the amino acids differ by 1 occurrence between various calmodulin sequences from distinct species, which shows that the types of amino acids in the various calmodulin sequences are conserved, whereas other data supports the conservation of the actual sequence.
Overall, the above data and literature citations indicate that the calmodulin protein is found in various and diverse species of eukaryotes and that such calmodulin proteins have a highly conserved primary amino acid sequence.\
It is important and interesting to note that although the sequences of calmodulin is conserved across various eukaryotic species, research suggests that there are immunological differences (Onek et al., 1992). For instance, polyclonal antibodies against spinach calmodulin will not recognize vertebrate or protozoan calmodulin proteins. This lack of cross recognition by species specific calmodulin antibodies suggests that differences do existence within the epitopes (specific sequences where antibodies recognize and bind) of calmodulin proteins from distinct species. Maybe the small differences between various calmodulin sequences across extremely distinct species exist within the epitope of the calmodulin protein. One paper indicates that the difference among epitope sequences of distinct species' calmodulin sequences may be an evolutionary advantage so that cells can identify foreign calmodulin proteins instead of mistakenly treating the foreign calmodulin as "self" (Onek et al., 1992).
Prokaryotes:
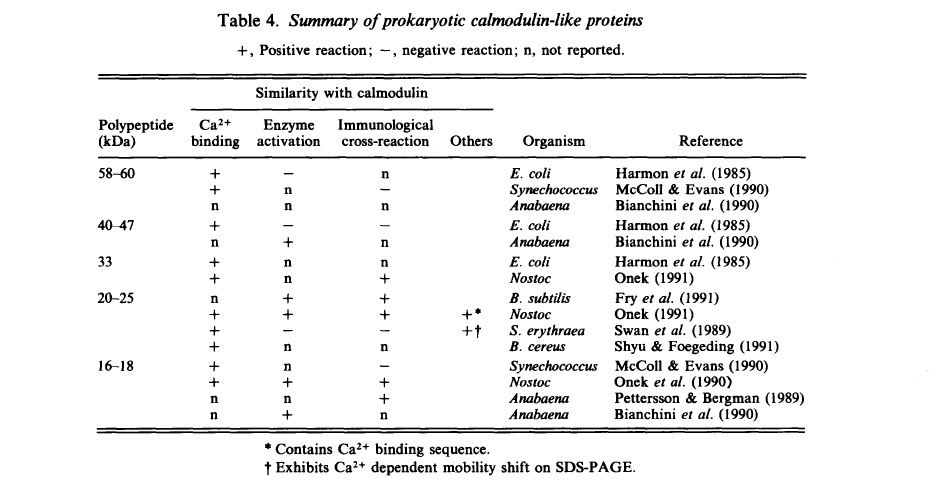
Initially researchers believed that calmodulin did not exist in bacteria (Klee et al., 1982), however in the past three decades evidence has surfaced suggesting the existence of proteins in many different species of bacteria that possess calmodulin-like properties. Researchers indicate that the difficulty in identifying calmodulin in bacteria is because of the stringent criteria for identification, which is based on the assumption that strong conservation of the calmodulin protein sequence extends to prokaryotes (Onek et al., 1992). The below table represents multiple prokaryotic proteins that show some form of similarity with the eukaryotic calmodulin protein.
Although evidence exists to suggest that some prokaryotic proteins exhibit calmodulin-like properties, evidence for true calmodulin is still not concrete. Yang, in another paper published in 2001, attempts to establish more concrete evidence for the existence of calmodulin within prokaryotes (Yang, 2001). Yang notes multiple proteins in prokaryotes, including E. coli, that show anywhere between 35-43% similarity to eukaryotic calmodulin seuqences using BLAST. BLAST alignments between different eukaryotic calmodulin sequences are above 55% similarity, indicating that these prokaryotic proteins may be part of an early branch of the calmodulin protein gene family because they share less similarity to later eukaryotic descendents (Yang, 2001). Therefore some sequence analysis suggests that prokaryotes possess the calmodulin protein, but most scientists agree that a deeper understanding of the role of calcium signaling is needed in order to fruther explore the possible existence of calmodulin.
Conclusions
Overall, an extensive literature search concerning the homology of the calmodulin protein indicated that calmodulin is found in most eukaryotes and its primary amino acid sequence is highly conserved among such eukaryotes. These two conclusions suggest that calmodulin is essentail for various life forms and essential for the proper functioning of life-sustaining pathways because calmodulin is found in most eukaryotes it must be conserved for a particular reason. The fucntion of calmodulin is essential for vairous pathways in almost all eukaryotes as noted in the previous webpage created concerning function and structure. The calmodulin function of calcium binding singal transducers is consistent throughout all eukaryotes and all eukaryotes rely on calcium as a intracellular second messanger (Yang, 2001), therefore the conservation of calmodulin and calmodulin sequence/structure throughout most eukaryotes is appropriate and necessary. These two conclusions also suggest that the primary amino acid seuqence has been conserved over time because it is extremely important for the function of the protein. This is also verified by the previous webpage, which indicates that the 4 calcium binding EF-hands along with the connecting string of amino acids are essential to proper functioning of the calmodulin protein. Further research indicates that the calcium binding hands are 4 separate and homolgous domains within each calmodulin protein and it is the primary amino acid sequence for each hand which is highly conserved to ensure proper functioning (Nojima, 1987). In terms of prokaryotes, nothing concrete can be declared for any of my research findings, but the suggestion of a previous ancestor preceding the modern eukaryotic calmodulin is a viable hypothesis for the proteins discovered in prokaryotes that share some calmodulin-like properties with the eukaryotic calmodulin.
Please direct questions or comments to karicheson@davidson.edu