THIS WEBSITE WAS PRODUCED AS AN ASSIGNMENT FOR AN UNDERGRADUATE COURSE AT DAVIDSON COLLEGE
Taq DNA Polymerase Orthologs
An ortholog is a gene that is common in two different species because it has evolved from a common ancestor. Taq DNA polymerase is trancribed by RNA translated from genes that are orthologous to genes in other species. These genes code for similar proteins, which are expressed in other species (Sadava et al., 2008). DNA polymerase is an enzyme that is present in all species because it is vital for DNA replication. The chart below compares amino acid sequences of DNA polymerases in Taq and other species:
Feature 1 ## # ## # # ## ######
T. aquaticus 559 RLHTRFNQT.[1].TATGRLSSSDPNLQNIP.[6].QRIRRAFIAE.[2].WLLVALDYSQIELRVLAHL.[1].GDENL 628
G. stearothermophilus 305 KVHTIFNQA.[1].TQTGRLSSTEPNLQNIP.[6].RKIRQAFVPS.[3].WLIFAADYSQIELRVLAHI.[1].EDDNL 375
X. laevis (African clawed frog) 806 KYGAILAQV.[3].GTITRRAVEPTWLTASN.[7].SELKAMVQVP.[2].YHLIGADVDSQELWIAAIL GEAHF 877
T. adhaerens 766 KYGAILPPV.[3].GTVTRRAVEATWLTASN.[7].SELKAMIRAP.[2].YHFVGADVDSQELWIAALY ADARF 837
D. melanogaster (fruit fly) 754 AYGAICPQV.[3].GTLTRRAMEPTWMTASN.[7].SELRSMVQAP.[2].YRLVGADVDSQELWIASVL GDAYA 825
M. brevicollis MX1 543 PIGAIIPMV.[3].GTVTRRAVEKTWMTASN.[7].SELKAMIQAP.[2].YKLVGADVDSQELWIASVI GDQRL 614
S. pombe (fission yeast) 627 GFGIILPCI.[3].GTVTRRAVENTWLTASN.[7].SELKAMIRAP.[2].YTFVGADVDSEELWIVALM GDSQF 698
Feature 1 # # # ## # # #
T. aquaticus 629 IRVF.[ 3].RDIHTETASWM.[11].MRRAAKTINFGVLYGMSAHRLSQEL.[ 4].EEAQAFIERYF.[22].VETLF 724
G. stearothermophilus 376 MEAF.[ 3].LDIHTKTAMDI.[11].MRRQAKAVNFGIVYGISDYGLAQNL.[ 4].KEAAEFIERYF.[22].VTTLL 471
X. laevis (African clawed frog) 878 AGIH.[17].TDLHSKTASTV.[ 2].SREHAKVFNYGRIYGAGQPFAERLL.[10].QAAEKAKQMYA.[65].ESQMF 1027
T. adhaerens 838 GRTH.[17].TDLHSKTAETV.[ 2].SRDQAKTFNYGRIYGAGLNYAQRLM.[10].DASDKAKILYA.[64].ESEMF 986
D. melanogaster (fruit fly) 826 CGEH.[17].SDMHSITAKAV.[ 2].SRDHAKVINYARIYGAGQLFAETLL.[10].EAKAKAMKMFS.[52].ESAMF 962
M. brevicollis MX1 615 GREH.[17].TDMHSMTAKIL.[ 2].NRDEAKIFNYGRIYGAGKAFAASLL.[10].EAQAKAEELYL.[18].ESHMF 717
S. pombe (fission yeast) 699 RLHG.[16].TDLHSKTAAIL.[ 2].SRDSAKVFNYGRLYGAGLKHTTLLL.[10].EAKELAKKLYA.[28].ESFVF 810
Feature 1 # # # # ####
T. aquaticus 725 GRRRYV.[14].ERMAFNMPVQGTAADLMKLAMVKLFPRLE.[4].RMLLQVHDELVLEAP.[4].EAVARLAKEVME 808
G. stearothermophilus 472 HRRRYL.[14].ERMAMNTPIQGSAADIIKKAMIDLNARLK.[6].HLLLQVHDELILEAP.[4].ERLCRLVPEVME 557
X. laevis (African clawed frog)1028 NKLETI.[28].ITSRVNWVVQSSAVDYLHLMLVAMKWLFE.[6].RFCISIHDEVRYLVH.[5].RAALALQITNLL 1128
T. adhaerens 987 NALEAI.[28].LPSRINWVVQSSAVDYLHLILVCMRWLFE.[6].RFCISIHDEVRYLVK.[5].RAALALQITNLL 1087
D. melanogaster (fruit fly) 963 NRLEEI.[31].LPTRINWVVQSGAVDFLHLMLVSMRWLMG.[3].RFCLSFHDELRYLVK.[5].KAALAMHITNLM 1063
M. brevicollis MX1 718 TSLEDI.[36].MTSRVNWVVQSSAVDFLHLMITSMDHFIR.[6].RLCITIHDEIRYIVA.[5].RVAYALHLTNLY 826
S. pombe (fission yeast) 811 NKLEAM.[29].MTSRVNWAIQSSAVDYLHLLLVSMNHLIK.[6].RLSLTVHDEVRYLSS.[5].RVAFALQVANLW 912
Feature 1
T. aquaticus 809 .[ 9].EVEVGIGEDWIS 829
G. stearothermophilus 558 .[ 9].KVDYHYGSTWYD 578
X. laevis (African clawed frog)1129 .[22].AVDIDKCLRKEV 1162
T. adhaerens 1088 .[22].AVDVDTVLRKFP 1121
D. melanogaster (fruit fly) 1064 .[22].SVEVDTVLRKEC 1097
M. brevicollis MX1 827 .[22].AVDVDTVMRKEV 860
S. pombe (fission yeast) 913 .[22].SVDIDHVLRKDV 946
Fig. 1. Conserved sequences of DNA polymerases in seven species. These species contain DNA polymerases with amino acid sequences that
are most closely related to the amino acid sequence of Taq DNA polymerase with conserved amino acids in red. The model organisms Drosophila
melanogaster and Schizosaccharomyces pombe (a yeast), are included in the comparison. (http://www.ncbi.nlm.nih.gov/)
Human DNA polymerases
There are fourteen DNA polymerases that have been identified in humans. There is debate about the roles of each polymerase enzyme and much is still unknown about them, though it is generally regarded that DNA polymerase δ is responsible for conventional DNA replication (Sadava et al., 2008). In their paper, Delarue et al. compare the structure of human DNA polymerase α, another human DNA polymerase, with various other DNA polymerase enzymes from different species to find conserved regions. Delarue divides all polymerases into three families (though there is evidence of five families): pol alpha's, pol beta's and pol I's, with human DNA polymerase α in the pol alpha category. He notes that pol alpha's and pol I's are highly conserved at three motifs (see fig. 2). Motif A contains an aspartate between an α-helix and a β-strand, motif B contains an α-helix with positive charges and motif C contins negative charges in a beta-turn-beta alignment. It is hypothesized that motifs A and B bind DNA, which is why they are so conserved among different species. (Delarue et al., 1990).
E. coli DNA polymerases
Five DNA polymerases have been identified in E. coli. It is DNA polymerase III that is responsible for conventional DNA replication, polymerase II that repairs DNA cross-links and polymerase I that is used in excision repair and Okazaki fragment processing (Albá, 2001). DNA polymerase I in E. coli contains regions that are highly conserved, namely the polymerizing region and six motifs in the 5’-3’ exonuclease region (Kim et al., 1995). Because of their similarities, both Taq polymerase and E. coli polymerase I are of the pol I family. Of the “big seven” model organisms, the functional sites of Taq DNA polymerase are most highly conserved in E. coli. The Klenow fragment of E. coli pol I, which is responsible for 5’-3’ polymerase activity shares 51% of its amino acids with the polymerizing region of Taq DNA polymerase (Kim et al., 1995). In addition, it shares six highly conserved motifs with ten conserved acidic residues. These sites exist at the 5’-3’ exonuclease regions of both enzymes. See fig. 2 for comparison.
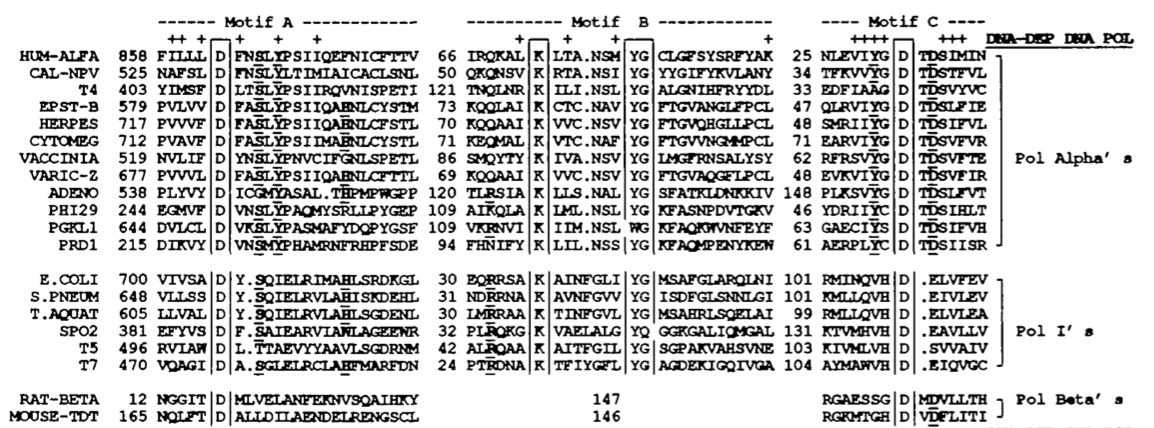
Fig. 2. Conserved sequences of DNA polymerases. Strictly conserved proteins are boxed and
highly conserved residues are bold and underlined. The polymerases represented are human
DNA polymerase α, A. californica nuclear polyherosis virus, phage T4, Epstein-Barr virus, cyto-
megalovirus, vaccinia virus, varicella-Zoster virus, cytomegalovirus, adenovirus, phage 29, yeast
plasmid PGKL1 and phage PRD1. Each of these are considered Pol Alpha’s, because of their
commonalities. The Pol I’s below also have much in common and are from E. coli, S. pneumoniae,
Taq, and bacteriophages SPO2, T5, and T7. The Pol Beta’s include rat polymerase β and mouse
terminal transferase. The motifs are N-terminal (Motif A), and C-terminal (Motifs B and C).
(Delarue et al., 1990) Permission Pending.
Overall Conservation
As explained on my favorite protein page, the structure of the polymerizing region of Taq polymerase is similar to an open hand, with "fingers" that recognize the dNTPs. The "palm" region of this hand is most highly conserved in all families of DNA polymerases, but the "fingers" and "thumb" domains vary extensively from family to family (Steitz, 1999). Fig. 3 shows a comparison of these domains from four different families.
Another conserved element is the mechanism by which DNA polymerase catalyzes each new phosphodiester bond. The reaction requires two Mg2+ ions to bind to conserved apartate residues in motifs A and C (see fig. 2). These residues are conserved throughout all families because they seem to be necessary for the reaction to occur (Albá, 2001). Both this mechanism and the "palm" region are highly conserved in DNA polymerases from all species, including the "big 7" genomic species. Figs. 1 and 2 show specific comparisons from five of the seven species. While Arabidopsis and C. elegans are not specifically sequenced, both contain DNA polymerases with the conserved divalent ion binding site and conserved "palm" region.
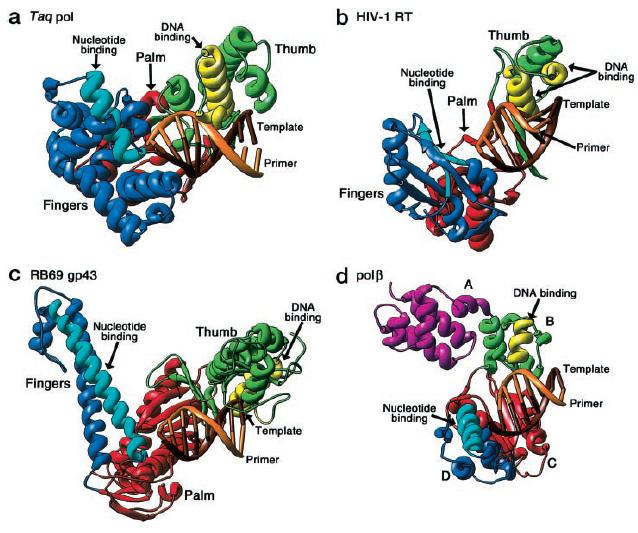
Fig. 3. Structural differences between families of DNA polymerases. Structures of Taq polymerase (a, pol I family),
HIV reverse transcriptase (b, unspecified family), RB69 gp43 (c, pol alpha family), and pol β (d, pol beta family). The
palm structures (red) are highly conserved whereas the thumb (green/yellow) and finger (blue/light blue) are not.
(Steitz, 1999) Permission Pending.
References
Albá, MM. Replicative DNA polymerases. Genome Biology 2001; 2: 3002.1-3002.4 http://www.biomedcentral.com/content/pdf/gb-2001-2-1-reviews3002.pdf
Delarue M, Poch O, Tordo N, Moras D, Argos P. An attempt to unify the structure of polymerases. Protein Engineering 1990; 3: 461-467. http://lorentz.dynstr.pasteur.fr/website/publi/delarue_proteng.pdf
Jung G, Leavitt MC, Jui-Cheng H, Ito J. Bacteriophage PRD1 DNA polymerase: Evolution of DNA polymerases. Proc. Natl. Acad. Sci.; 84: 8287-8291. http://www.jstor.org/stable/30447
Kim Y, Eom SH, Wang J, Lee D, Suh SW, Steitz TA. Crystal Structure of Thermus aquaticus DNA polymerase. Nature 1995; 376: 612-616. http://www.nature.com/nature/journal/v376/n6541/pdf/376612a0.pdf
Sadava H, Heller HG, Orians GH, Purves WK, Hillis DM. Life: The Science of Biology. New York (NY): WH Freeman and Company; 2008. 1251 p.
Steitz, TA. DNA Polymerases: Structural Diversity and Common Mechanisms. Journal of Biological Chemistry 1999; 274: 17395-17398. http://www.jbc.org/content/274/25/17395.full.pdf+html
For more information about Taq DNA Polymerase, visit its pages at GenScript and EBI
Molecular Biology Homepage - Davidson College Homepage - My Homepage
Questions and comments can be directed to ststreb@davidson.edu
