THIS WEBSITE WAS PRODUCED AS AN ASSIGNMENT FOR AN UNDERGRADUATE COURSE AT DAVIDSON COLLEGE
Taq DNA Polymerase

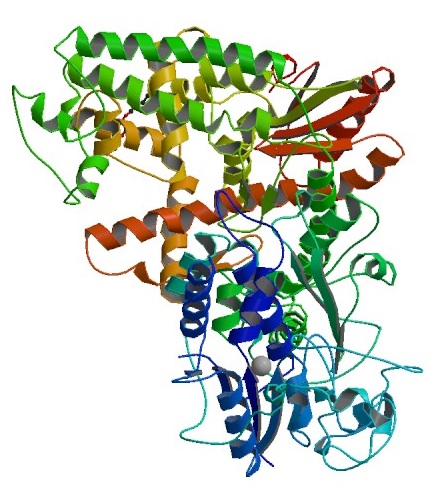
Fig. 1. Crystal Structure of Taq DNA Polymerase (Protein Data Bank)
Background
Taq DNA Polymerase is a thermostable DNA polymerase present in Thermus aquaticus, a thermophile. Thomas D. Brock isolated T. aquaticus DNA polymerase from the species in a hot spring at Yellowstone National Park (Sadava et al., 2008). Due to its resistance to heat denaturation, Taq DNA polymerase has a host of uses in biotechnology, including being the DNA polymerase of choice for the Polymerase Chain Reaction (Erlich, 1991).
Function
The function of a DNA polymerase enzyme is to facilitate the synthesis of DNA molecules from monomeric deoxyribonucleotide triphosphate subunits (Burrell, 1993). This process is essential for all organisms, and therefore all organisms contain some form of DNA polymerase. Some DNA polymerases have exonuclease activity, meaning they can cleave nucleotides from a DNA strand. This is useful in proofreading and DNA repair. Taq DNA polymerase does not have 3’ to 5’ exonuclease activity, but it does have 5’ to 3’ exonuclease activity (Kim et al., 1995).
Taq DNA Polymerase has an optimum temperature of 80°C (Chien et al., 1976). According to Lawyer et al., it has maximal activity when in a solution of 2-4 mM MgCl2 and 10-55 mM KCl. Mg2+ satisfies the requirement of a divalent cation cofactor, which is necessary for catalysis of DNA synthesis (Chien et al., 1976). Also necessary for catalysis are a primer, dNTPs, and a DNA template (Sadava et al., 2008).
Structure
The Taq DNA Polymerase gene is 2499 nucleotides long. It codes for a protein that is 832 amino acids in length and has a molecular weight of 94 kD. It is composed of one polypeptide chain (Burrell, 1993).
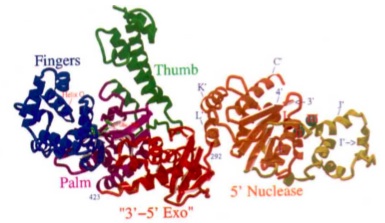
Fig. 2. Structure of Taq DNA Polymerase with the “open hand” structures of the polymerization region,
the 3’ to 5’ exonuclease region (nonfuctional), and the 5’ nuclease region labeled (Kim et al., 1995).
Polymerizing Region
The polymerizing region of a DNA polymerase is shaped like an “open hand” whose “fingers” are able to recognize the four deoxyribonucleotide bases (Sadava et al., 2008). This region of Taq DNA Polymerase is located at the enzyme’s carboxyl terminus. According to Kim et al., this region is “nearly identical” to the Klenow fragment, a protein fragment of DNA polymerase I from E. Coli that contains the polymerizing region. The “open hand” of Taq DNA Polymerase (see Fig. 2) wraps around a DNA strand, first binding to a primer. It undergoes a conformational change, the "finger" domain rotating inward by 46° to add nucleotides. Evidence of these distinct conformations, called "closed" and "open" respectively, is provided by Li et al.
Central Region
The central region of Taq DNA Polymerase is the region that corresponds to the 3’ to 5’ exonuclease domain on the Klenow fragment. According to Kim et al., this region “differs extensively” from the Klenow fragment. On the Taq enzyme, four loops of lengths between 8 and 27 residues have been deleted. Also, all four of the carboxylates, which are necessary for metal-ion binding, and thus necessary for functionality of the region, are not present. These deletions are consistent with the fact that Taq DNA Polymerase cannot act as a nuclease in the 3’ to 5’ direction.
Exonuclease Region
The 5’ to 3’ exonuclease domain of Taq DNA Polymerase can function independently after proteolytic removal from the rest of the protein. This region has a cleft, inside of which are carboxylates which bind divalent metal ions. Additionally it should be noted that there are six highly conserved sequence motifs in this region from E. Coli DNA polymerase I, including the spherical structure visible in the both diagrams of Fig. 3 (Kim et al., 1995).
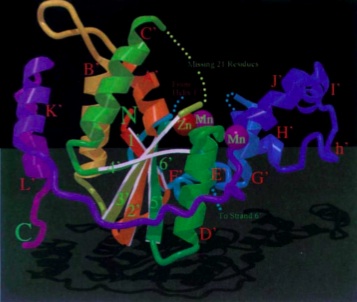
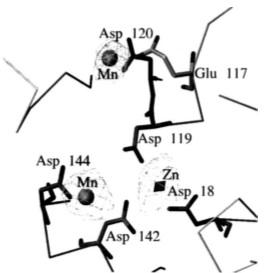
Fig. 3. 5’ exonuclease region of Taq DNA Polymerase. Left: ribbon diagram of the
5’ nuclease
region.
Right: metal ion binding sites on the 5’ nuclease region (Kim et al., 1995)
Thermal Stability and Structure
Compared to E. Coli DNA Polymerase I, Taq DNA Polymerase contains four additional hydrogen bonds, two formerly hydrophilic salt bridges are hydrophobic, the ratio of leucine to isoleucine is 4.4-fold higher and the ratio of arginine to lysine is 1.3-fold higher. According to Kim et al., this is not definitive proof of thermal stability, but this information is consistent with the thermal stability of Taq DNA Polymerase.
References
Burrell MM, editor. Enzymes of Molecular Biology. Totowa (NJ): Humana Press; 1993. 370 p.
Chien A, Edgar DB, Trela JM. Deoxyribonucleic Acid Polymerase from the Extreme Thermophile Thermus aquaticus. Journal of Bacteriology 1976; 127: 1550-1557.
Erlich HA, editor. PCR Technology: Principles and Applications for DNA Amplification. New York (NY): WH Freeman and Company; 1992. 246 p.
Kim Y, Eom SH, Wang J, Lee D, Suh SW, Steitz TA. Crystal Structure of Thermus aquaticus DNA polymerase. Nature 1995; 376: 612-616.
Lawyer FC, Stoffel S, Saiki RK, Chang S, Landre PA, Abramson RD, Gelfand, DH. High-level expression, purification, and enzymatic characterization of full-length Thermus aquaticus DNA polymerase and a truncated form deficient in 5’ to 3’ exonuclease activity. Genome Research 1993; 2: 275-287.
Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. The EMBO Journal 1998; 17: 7514-7525.
Sadava H, Heller HG, Orians GH, Purves WK, Hillis DM. Life: The Science of Biology. New York (NY): WH Freeman and Company; 2008. 1251 p.
For more information about Taq DNA Polymerase, visit its pages at GenScript and EBI
Molecular Biology Homepage - Davidson College Homepage - My Homepage
Questions and comments can be directed to ststreb@davidson.edu