*This website was produced as an assignment for
an undergratuate course at Davidson College.*
Gal4
from Saccharomyces cerevisiae
The Story of Orthologs...
What is an Ortholog?
An ortholog is a protein with high homology to a
proteins found in another species. As species diverge, both new
species carry many of the same genes (Figure 1). These genes will
often evolve differently forming two similar, but not identical
genes. These two genes then encode for the proteins that are
considered orthologs because they have high similarity but are found in
different species.
|
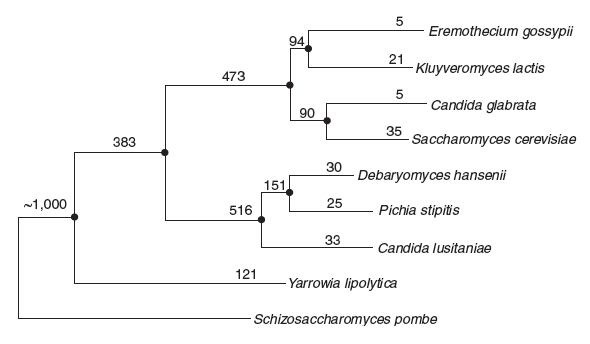
Figure 1. Phylogenetic
tree of seven sequenced hemiascomycetous yeast genomes based on
multiple alignment of 94 single-copy genes conserved in 26 tasonomic
groups (see Methods). Numbers next to each branch correspond to the
number of families (clusters) specific to a genome or group of genomes
leading to this node. (Figure Reproduced with Permission, 1)
|
Background on Gal4 Orthologss
Orthologs of the Gal4 protein were not found in any other
species included in the Big Seven Genomic Species (consisting of human,
mouse, C. elegans, Drosophila, Arabidopsis, yeasts, and E. coli)
except for yeast contained Gal4 orthologs. An extensive
literature search was performed as well as using BLAST, DB Phylome, and
Ortho DB databases (BLAST Results, DB Phylome Results, and Ortho DB Results).
Of the 15 genes from 10 species found in the Ortho DB database search,
it was found that only a handful had been characterized, and further
only a few had been extensively characterized.
The literature search confirmed that the Gal4 family
of transcription factors is in fact a fungal specific family
(2-4). Interestingly, yeasts are eukaryotic organisms and are
often used as a model due to similarities to human cells. The
Gal4 protein, however, is not one of the similarities. This
implies that this family of proteins developed after fungi diverged
from other eukaryotes.
The remainder of this page will review the homology
seen between Gal4 and one of the other most extensively characterized
orthologs, the Lac9 protein in Kluyveromyces lactis.
A comparison of these orthologs has proven useful for identifying
critical sequences in the function of Gal4 and Lac9 proteins. No
useful literature for the other orthologs reported in the Ortho DB
search was found.
A Comparison: Gal4 vs. Lac9
The Gal4 protein is necessary for harvesting galactose from the major food source of S. cerevisiae, melibiose (Gal4) (2). A different strain of yeast, K. lactis,
is found in milk where its primary food source is lactose (2). In
order to harvest galactose from lactose, it uses a slightly different
set of proteins than does S. cerevisiae. Ultimately both species obtain galactose from their respective environments (2).
The two species contained a common ancient ancestor,
making it likely that they share many of the same genes (2). In
fact high conservation between the two species is seen (Figure
1). Often conserved sequences are the most highly studied
sequences because a useful way in understanding functions of recently
identified proteins comes from comparison with proteins in other
organisms that are “similar, yet divergent in both sequence and
function” (3-4). In this case, both strains of yeast must
accomplish similar tasks: obtaining galactose from a food source.
It is probably that by studying similarities between the proteins used
in the two yeasts, we can obtain significant information regarding the
proteins.
The Gal4 function has been described previously as necessary for regulation of galactose metabolism (Gal4). In the similar yeast strain, K. lactis, this metabolism is regulated by a Gal4 ortholog, the Lac9 protein (3). Upon analysis of the Lac9 protein in K. lactis, Salmerom et al., determined that there is significant functional similarity between the proteins encoded by the GAL4 and LAC9
genes, however the actual amino acid sequence is quite divergent
(2). Both proteins bind DNA as a “homodimer to specific upstream
activating sequences (UASG) in the GAL promoters” (2). However the Gal4
and Lac9 proteins contain only about 30% of the same amino acid
sequence (2-3).
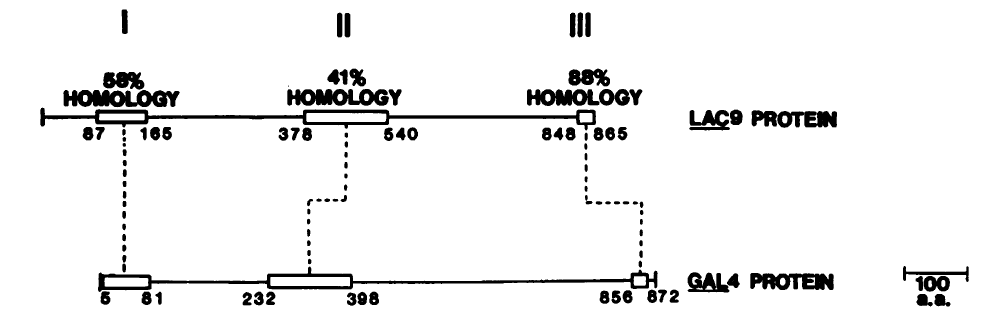
|
Figure 2. Regions of homology (open boxes) between LAC9 and GAL4 Proteins. Thin lines represent regions possessing no more than 17% homology between the two proteins. (Permission Pending, 3)
|
The homology between the Gal4 and Lac9 proteins
can be seen in three regions (Figure 2). Region I contains
a series of 76 amino acids with high homology (3). This region,
which is located on the N-terminus, has been shown to be required for
nuclear localization (3). Further it has been suggested that this
region is also necessary for DNA-binding as well, suggesting that the
two proteins have similar DNA-binding domains (4). In both
studies the researchers were placing functions on the conserved
sequences that are common to the two proteins. Because both
proteins must facilitate similar tasks within the cell, it makes since
that such functions would be contained within the conserved sequences.
For instance both act as transcription factors and therefore need to
localize to the nucleus. Without a correct signal sequence, this
would not be accomplished and the protein would not function.
Both proteins also need to bind DNA, and are expected to do so in a
similar manner. There it is not surprising that the conserved
regions of the protein are important for the common functions of the
two proteins.
Region II and Region III were not completely
described, but possible reasons for the conservation were
suggested. Region II, located close to the middle of both
proteins, could play a role in oligomerization or other interactions
that are common between the two proteins (3). Others suggested
that this region might interact with negative regulators, but also
state that because of its size it probably has more than one function
(4). Region III is a “short (18 amino acid), but almost
completely conserved, region located within this C-terminal area”
(3). Salmerom et al., suggest that this must be another
functional domain because little homology is seen surrounding it,
implying these 18 amino acids were conserved for a specific reason
(3). They however did not state what function they thought
would be carried out by the sequence. Another study suggested
other functions that are most likely contained within one of the
conserved regions: repression, subunit interation, and transcriptional
activation (4). It is possible that these functions also
correspond to one of the regions described.
In the end, the two proteins that contain much
similarity do have many differences. Due to the conservation of
certain sequences, it is expected that these contain critical
functional domains of the proteins (3-4).
Significance of This Orthology in Other Species
The lack of Gal4 orthologs in other species,
specifically Drosophila and mammals, provide many useful tools.
As probably one of the most widely characterized eukaryotic
transcription factors, many tool using Gal4 have been developed
(5). It is often used to identify protein-DNA and protein-protein
interactions using yeast one-hybrid and yeast two-hybrid screens
respectively. Further, when transfected into cells other than
yeast, the properties of Gal4 have allowed researchers to identify
various transcriptional activators among other things (5). Gal4
has become a very useful tool.
References
(1) Jeffries
TW, Grigoriev IV, Grimwood J, Laplaza JM, Aerts A, Salamov A, Schmutz
J, Lindquist E, Dehal P, Shapiro H, Jin Y, Passoth V, Richardson PM.
Genome sequence of the lignocellulose-bioconverting and
xylose-fermenting yeast pichia stipitis. Nat Biotechnol 2007
MAR;25(3):319-26.
(2) Rubio-Texeira
M. A comparative analysis of the GAL genetic switch between
not-so-distant cousins: Saccharomyces cerevisiae versus kluyveromyces
lactis. FEMS Yeast Res 2005 DEC;5(12):1115-28.
(3) Salmeron
JM, Johnston SA. Analysis of the kluyveromyces-lactis positive
regulatory gene Lac9 reveals functional homology to, but sequence
divergence from, the saccharomyces-cerevisiae Gal4 gene. Nucleic Acids
Res 1986 OCT 10;14(19):7767-81.
(4) Wray
LV, Witte MM, Dickson RC, Riley MI. Characterization of a positive
regulatory gene, Lac9, that controls induction of the lactose-galactose
regulon of kluyveromyces-lactis - structural and
functional-relationships to Gal4 of saccharomyces-cerevisiae. Mol Cell
Biol 1987 MAR;7(3):1111-21.
(5) Sadowski I. Uses for GAL4 expression in mammalian cells. Genet Eng (NY) 1995 1995;17:119-48.
Matt's Homepage
Molecular Biology Homepage
Davidson College Homepage
Please direct questions or comments to my email masurdel@davidson.edu


