*This website was produced as an assignment for an undergraduate course at Davidson College.*
ATP Synthase Function
ATP, adenosine triphosphate, provides the energy for many cellular processes, including active transport of molecules across cell membranes, motility, and metabolism. ATP synthase functions to make ATP for the cell to consume (von Ballmoos, Wiedenmann, Dimroth, 2009). ATP synthase enzymes are found in all organisms that undergo aerobic respiration and are located where cellular respiration takes place: the cell membrane in prokaryotes, the mitochondrial inner membrane in eukaryotes, and the chloroplast thylakoid membrane. In all cases, ATP synthases harness the electrochemical energy from a transmembrane proton or sodium (Na+) gradient to combine ADP and inorganic phosphate (Pi) to make ATP (Ackerman, Tzagoloff, 2005). In eukaryotes, the proton pumps of the mitochondrial electron transport chain create a proton gradient, in which the pH is lower (there is a greater H+ concentration) in the intermembrane space (P side) than in the mitochondrial matrix (N side) (Nelson, Cox, 2008).
ATP Synthase Structure

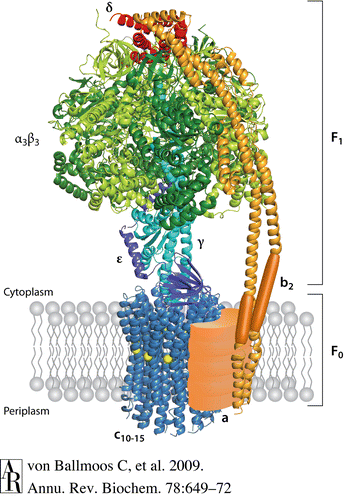
In broad terms, ATP synthases have two functional domains: an intramembrane, hydrophobic region called F0 and an N side, hydrophilic region called F1. Protons flow down their concentration gradient from the P side to the N side of the mitochondrial inner membrane through the proton channel located in the F0 portion of the ATP synthase complex, which generates energy that the F1 domain uses to catalyze the formation of ATP (Ackerman, Tzagoloff, 2005).
Escherichia coli is a model organism with respect to ATP synthase structure and has a quaternary structure composed of eight subunits. The F1 domain has five subunits, α, β, γ, δ, and ε, in the proportion α3β3γδε. The F0 portion of ATP synthase consists of three subunits, a, b, and c, and has one a component, two b components, and between 10 and 14 c components. The circular “head” portion of the ATP synthase complex, which encompasses the three sites for ATP catalysis, is a hexamer that consists of an alternating pattern of three α and three β subunits. The α and β subunits have an N-terminal β-barrel, a nucleotide binding site, and a C-terminal α-helix. Connected to the center of this “head” complex is the central stalk, which is comprised of one γ and one ε subunit. The γ subunit consists of two α-helices arranged in a coiled-coil configuration. Attached to the side of the “head” for stabilization is the peripheral stalk, which is one δ subunit on top of two b subunits. The membrane-embedded portion of the F0 domain contains one a subunit right next to a circular ring of c subunits that surrounds and interacts with the central stalk. The proton channel in the F0 domain is in between the a and c subunits (Ackerman, Tzagoloff, 2005).
The ring of c subunits and the F1 central stalk begin to turn in 120° increments when protons flow down their concentration gradient through the proton channel in F0 (Ackerman, Tzagoloff, 2005; von Ballmoos, Wiedenmann, Dimroth, 2009). The “head” portion of F1 does not rotate and is kept stagnant by the peripheral stalk. There are three catalytic, nucleotide-binding sites, one on each β subunit at the point where it meets the preceding α subunit. Each β subunit spontaneously and sequentially transforms into one of three conformations that have different ADP and ATP binding affinities: MgADP (βDP conformation, which binds ADP and Pi), MgATP (βTP conformation, which has a high binding affinity for ATP), and empty (βE conformation, which has a low binding affinity for ATP) (Ackerman, Tzagoloff, 2005; von Ballmoos, Wiedenmann, Dimroth, 2009). With each 120° turn of the central stalk, the γ subunit touches a β subunit in the βTP conformation and causes it to release its molecule of ATP and assume the βE conformation (Nelson, Cox, 2008). Each β subunit will assume all three conformations in a given rotational cycle, and at any given time each of the three catalytic sites is in a different conformation (von Ballmoos, Wiedenmann, Dimroth, 2009; Nelson, Cox, 2008). Thus, for each complete 360° turn of the central stalk, three molecules of ATP are synthesized, and it can rotate 700 times per second (von Ballmoos, Wiedenmann, Dimroth, 2009; Nelson, Cox, 2008)!
Only a few amino acids in ATP synthase’s primary structure have been discovered to be necessary for the enzyme’s function. An intramembrane, positively-charged arginine residue at E. coli position 210 in the F0 domain is highly conserved across species, and a mutation in this amino acid completely disables the E. coli ATP synthase enzyme. Arginine’s positive charge may help to repel the positively-charged H+ ions, which promotes their discharge from their c ring binding site to the N side of the mitochondrial inner membrane. Three amino acids embedded in the inner membrane near the surface on the P side—glycine at position 218 (G218), glutamic acid at position 219 (E219), and histidine at position 245 (H245)—seem to be important for trapping environmental protons for ATP synthase to use as a source of electrochemical energy. Certain mutations in one of these three amino acids can cause complete loss of enzyme function, specifically a change from glutamic acid to leucine or cysteine and a change from histidine to serine, cysteine, leucine, or tyrosine. A second mutation in either of the other two amino acid residues in this triad can correct the effect of one of these loss-of-function mutations, however, and restore ATP synthase function (von Ballmoos, Wiedenmann, Dimroth, 2009).
For more information:
ATP Synthase in Protein Data Bank
Works Cited
Ackerman SH, Tzagoloff A. 2005. Function, structure, and biogenesis of mitochondrial ATP synthase. Progress in Nucleic Acid Research and Molecular Biology 80: 95-133.
Goodsell DS. ATP synthase. [Internet]. 2005 Dec [cited 2010 Feb 16]. RCSB Protein Data Bank. Available from: http://www.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/pdb72_1.html
Nelson DL, Cox MM. 2008. Lehninger principles of biochemistry. 5th ed. New York (NY): W.H. Freeman and Company; p. 723-732.
von Ballmoos C, Wiedenmann A, Dimroth P. 2009. Essentials for ATP synthesis by F1F0 ATP synthases. Annual Review of Biochemistry 78: 649-672.