*This web page was produced as an assignment for an undergraduate course at Davidson College. *
Purpose
The purpose of this website will be to explore the various orthologs of the protein that Arylalkylamine N-Acetyltransferase (hereafter referred to as AANAT). An ortholog is a gene that has evolved, through speciation events, from a common ancestral gene and is found in a number of different species. As I already mentioned in the first website, the main function of AANAT in vertebrates is the catalysis of the limiting step in the production of melatonin, through the acetylation of serotonin. However, the AANAT found in vertebrates is only an ortholog of a protein found in bacteria that has distinct structural and functional differences (Klein, 2007).
.
The Beginning

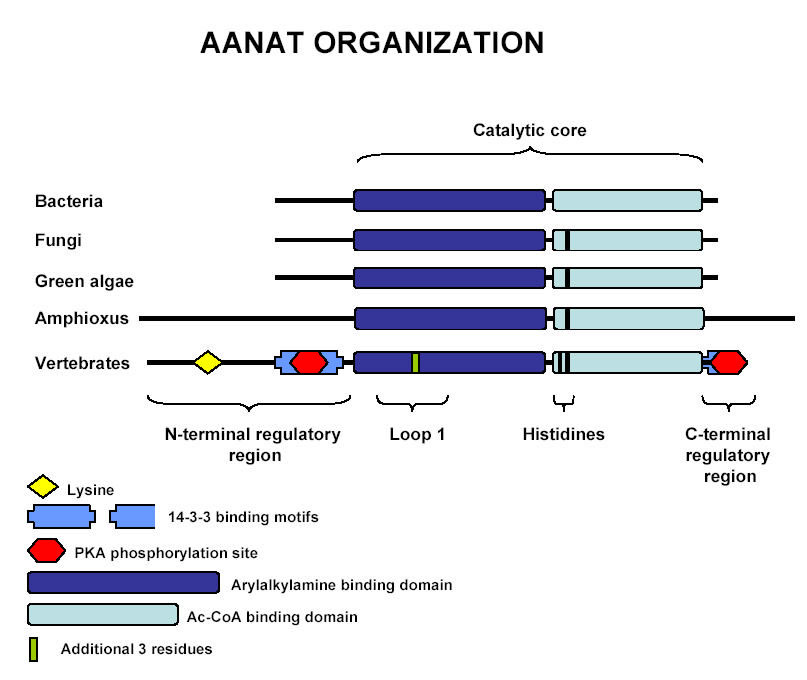
Figure 1. Schematic Representation of the AANAT orthologs in bacteria, fungi, green algae, amphioxus, and vertebrates. Picture obtained Coon and Klein, 2006. Permission Pending.
If we wanted to classify AANAT and all its orthologs, they would be placed in the GNAT superfamily. The GNAT superfamily is a family of acetyltransferases that all have Acetyl Coenzyme A binding site that is part of conserved catalytic core (Figure 1.) However, the AANAT orthologs found in different species also have a range of different substrates that each can acetylate (Coon and Klein, 2006). On the one hand vertebrate AANATs are highly selective for arylalkylamines, such as serotonin and dopamine and on the other hand bacteria’s' and fungi's AANATAs are less selective and have a wider array of substrates, for instance the yeast homolog can acetylate amines and polyamines that the vertebrate AANATs are not capable of acetylating (Coon and Klein, 2006). The existence of such a wider range of substrates for bacteria and fungi and also because it has been shown that the characteristic daily changes in the activity of AANATs, as seen in vertebrates, is not seen in bacteria and fungi points to a unrelated function of AANATs in these organisms. More specifically, scientists speculate that AANATs in bacteria and fungi serve an alkylamine detoxifying purposes in the organisms’ intracellular space (Klein, 2006; Coon and Klein, 2006).
The Expansion
Figure 2. Hypothetical transfer routes (doted lines) of the AANAT ortholog through Horizontal Gene Transfer starting from gram-positive bacteria. Picture obtained from Coon and Kein, 2006. Permission Pending.
As mentioned in Coon and Klein's review paper , AANATs are only found in a certain number of organisms, namely gram-positive bacteria, fungi, unicellular algae, cephalochordates and vertebrates but it is not found in other major genomes, such as those of higher plants, nematodes and anthropods. The most commonly proposed reasoning behind this pattern of expression of the AANATs orthologs in nature is that there were three different events of horizontal gene transfer, which involved the transfer of ancestral ortholog most probably found in bacteria. Apart from the characteristic type of evolution of this gene, the absence of introns in the AANATs of the fungi, algae, and cephalochordata, resembling bacterial genomes, is also supportive of the horizontal gene transfer theory. However, it is also possible that AANAT orthologs are present in other species' genomes but are unidentifiable because of extensive divergence from the ancestral bacterial original or even more likely became lost from other species' lineages (2006).
The Divergence
Figure 3. Alignment of AANAT orthologs (amino acid position 90-164) from a range of organisms, using CLUSTALW. Shaded amino acids are conserved. Picture adapted from Coon and Klein, 2006. Permission Pending.
As I mentioned in the beginning, the orthologs of AANAT while maintaining their acetyl transferase abilities also present certain structural differences that mirror functional differences. Unlike the bacterial version of AANAT, every other version of the protein has a conserved histidine molecule located between the Loop 2 and Motif A domains of the protein (Figure 3). Also, in the vertebrate lineage there is an additional histidine close to the original residue (Figure 3). Taking into consideration the fact that the histidine residues play a crucial role in the catalytic activity of the enzyme, scientists have suggested that these differences in histidine residue conservation explain the significant differences of enzymatic efficiency of AANATs from different lineages; the yeast ortholog is reportedly a thousand times less efficient than the vertebrate ones (Cook and Klein, 2006).
Figure 4. Alignment of AANAT orthologs (amino acid position 1-89) from a range of organisms, using CLUSTALW. Shaded amino acids are conserved. Picture adapted from Coon and Klein, 2006. Permission Pending.
One crucial functional difference between vertebrate and the rest of the AANAT orthologs is, as I mentioned above, the rhythmic changes that occur in vertebrates throughout the day with a peak in activity seen during the night (Klein, 2006). This rhythmic feature is probably facilitated by the existence of a number of regulatory regions (Figure 1), which control phosphorylation/dephosphorylation and mediation/inhibition of proteasomal proteolysis, that are only present in the vertebrate versions of AANAT (Coon and Klein, 2006). Such regulatory regions include PKA phosphorylation sites (Figure 4) which both control phosphorylation but also promotes binding to the 14-3-3 proteins that stabilize and protect the enzyme while at the same a conserved lysine residue (Figure 4) is thought to mediate proteolysis(Cook and Klein, 2006). In addtion to the regulatory regions, there is also divergence in the catalytic domain of the AANAT orthologs. The vertebrate ortholog has an extra three amino acids located in the Loop 1 region (CPL, Figure 4) that have been associated with the loop's flexibility and specificity. As Klein mentioned in his review, these structural divergence could explain how "the highly active and precisely regulated "Timeyme" may have evolved from a relatively sluggish detoxifying enzyme"(2006)
Works Cited
Ilias Theodorou's Home Page Molecular Biology Home Page
For any questions or comments please email iltheodorou@davidson.edu