*This website was produced as an assignment for an undergraduate course at Davidson College.*
Orthology is an important component of evolutionary biology and modern genomics. Orthologs are genes in different species that have evolved from a common ancestral gene through speciation. With the availability of complete genomes of diverse organisms, scientists can conduct comparative analyses to gain a deeper understanding of general trends in the evolution of genomic complexity and lineage-specific adaptations (Koonin, 2005). Soon, it may even be possible to reconstruct the evolutionary history of each gene. Some of the evolutionary events that we must take into account include speciation (vertical descent), gene duplication, gene loss, horizontal gene transfer, and gene rearrangements (Koonin, 2005).
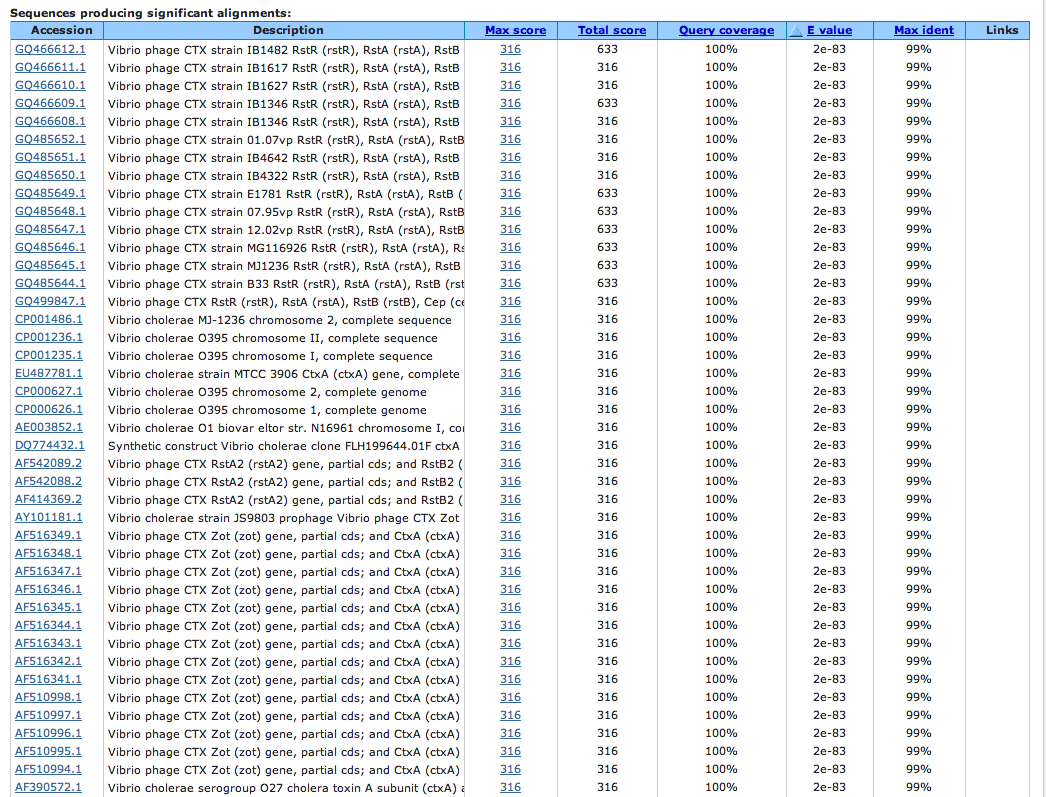
After an extensive literature and database search, orthologs of cholera toxin were found in only one of the big seven genomic species, Escherichia coli, and none in the remaining species (human, mouse, C. elegans, Drosophila, Arabidopsis, yeast). An NCBI BLAST search was conducted (175 nt query) and confirms that cholera toxin is a bacterial-specific protein with no orthologs in any of the big seven species except for E. coli. It is interesting to note that the BLAST query search resulted in sequences exclusively within Vibrio phage, various Vibrio cholerae isolates, and synthetic constructs of V. cholerae clones (Figure 1).
The ortholog of cholera toxin, E. coli heat-labile toxin, is a member of the ADP-ribosylation toxin family and causes watery diarrhea similar to that of cholera toxin. The remainder of this webpage will describe the homology between E. coli heat-labile toxin and cholera toxin. In addition, I will describe phylogenetic relationships and variation among Vibrio cholerae isolates as well as the evolution of new variants of V. cholerae.

Obtained from NCBI Blast. Permission pending.
Figure 1. BLAST query search of the first 175 nucleotides of the cholera toxin sequence. Note that the query results exclusively list different variants of Vibrio cholerae isolates or Vibrio phage. There were no sequence homologies listed from any of the big seven genomic species.
Cholera toxin and E. coli heat-labile toxin are structurally and functionally similar enterotoxins (Dallas & Falkow, 1980). Both enterotoxins are released in the intestine and cause elevated cyclic AMP levels due to the NAD-dependent ADP ribosylation of membrane proteins. This sustained increase of cAMP causes activation of intestinal sodium pumps, resulting in significant electrolyte and water loss, and sometimes life-threatening dehydration (Peterson & Ochoa, 1989). Moreover, each toxin is structurally composed of an enzymatic A chain as well as a targeting B chain. Nucleotide sequence comparisons of the two toxins suggest that the two proteins may have evolved from a common ancestor through a series of single selective base changes (Yamamoto et. al., 1984). Both cholera toxin (CT) and heat-labile enterotoxin (LT) share the same number of nucleotides and the same number of amino acids (Figure 2). However, there is still a significant amount of heterogeneity in the catalytic fragment A1 in both amino acid number and amino acid sequence (Figure 3). This seems to suggest that both LT and CT genes evolved from a common ancestor while each retaining their own A1 fragment.

Obtained from Yamamoto et. al., 1984. Permission pending.
Figure 2. Sequence comparison of E. coli heat-labile toxin and cholera toxin A genes. The dots signify any nucleotides or amino acid residues of cholera toxin that are identical to those of heat-labile toxin. Note that there is approximately 80% homology between the two toxins.
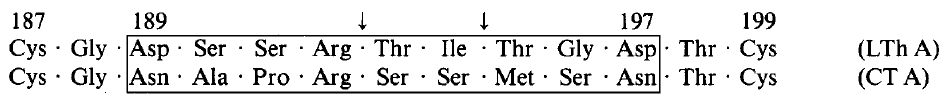
Obtained from Yamamoto et. al., 1984. Permission pending.
Figure 3. Divergent region in subunit A of E. coli and V. cholerae. This portion of the A1 fragment differs quite a bit between LT and CT. There is a unique random base change pattern. Yamamoto et. al. speculate that the divergence is a result of adaptations contributing to distinct enzymes, which may explain the evolutionary separation.
As seen in figure 2, LT and CT genes share the same number of nucleotides. The homology between LT and CT at the nucleotide level is 77.9%, while the homology at the amino acid level is 78.8% (Figure 4). The resemblance between the two toxins is confirmed by the similar G + C content; G + C for LT is 38.2% while CT is 38.5% (Yamamoto et. al., 1984).
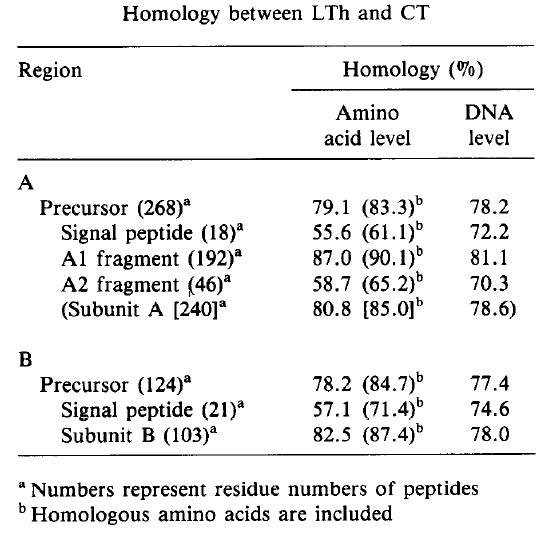
Obtained from Yamamoto et. al., 1984. Permission pending.
Figure 4. Percent homology between Escherichia coli heat-labile toxin and cholera toxin. There is a great deal of homology between the two toxins at both the amino acid level and the DNA level.
Cholera toxin is an 84,000-molecular weight protein composed of one A subunit (27,000 MW) and five B subunits (11,600 MW) (Mekalanos et. al., 1983). It is a heat-labile enterotoxin secreted by growing vibrios; the Gram-negative bacterium Vibrio cholerae is responsible for seven world pandemics of the diarrheal disease. Many members of the family Enterobacteriaceae are facultatively anaerobic, Gram-negative rods that are opportunistic pathogens similar to cholera toxin. The Enterobacteriaceae family is in fact responsible for approximately 50% of nosocomial infections in the United States (Paradis et. al., 2005). Cholera toxin is indeed phylogenetically similar to many of the members of the Enterbacteriaceae family (i.e. Shigella dysenteriae, Salmonella virchow, etc.). In many cases, scientists group cholera toxin along with the Enterobacteriaceae family, yet it is distinguished as an outgroup (Figure 5).
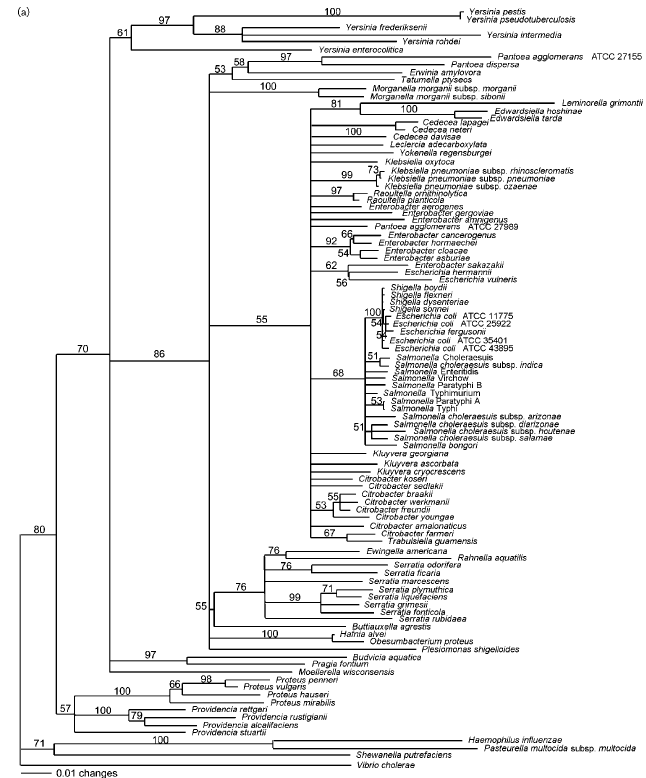
Obtained from Paradis et. al., 2005. Permission pending.
Figure 5. Phylogenetic tree of the Enterobacteriaceae based on genes encoding elongation factor Tu. Note that Vibrio cholerae is positioned at the very bottom of the tree. V. cholerae is used as an outgroup because it does not belong to the family Enterobacteriaceae; however V. cholerae is phylogenetically close to the family and is relevant in this case. Values on the branches indicate the occurence (%) of the branching in 1000 subsets.
Cholera pandemics have been caused by various serogroups (group of bacteria containing a common antigen) of Vibrio cholerae. New strains and serogroups have coexisted and have been responsible for larger outbreaks (Danin-Poleg et. al., 2007). The various isolates of V. cholerae have been divided into three major groups: O1, O139, and non-O1/non-O139. Environmental strains from the non-O1/non-O139 group have been cited as a potential reservoir for new emerging strains (Danin-Poleg et. al., 2007). It is important to understand that the development of new strains may be through horizontal gene transfer events (Stine et. al., 2000). In order to understand the epidemiology of pandemic strains and to monitor routes of contamination, genome-based bacterial identification and typing was used to study 32 isolates of V. cholerae (Figure 6). There exists a distinct separation between clinical and environmental groups of isolates. Based on the polymorphisms present between strains, there is speculation that the polymorphisms may have a functional role affecting gene regulation and the expressed protein (Danin-Poleg et. al., 2007). It is possible that these changes in the regulatory regions of the bacterial species contribute to the variation in gene expression and to the dynamic responses towards environmental changes (Danin-Poleg et. al., 2007).
(a)
(b)
Obtained from Danin-Poleg et. al., 2007. Permission pending.
Figure 6. (a) Sequence alignment of Vibrio cholerae isolates. Single nucleotide polymorphisms (SNPs) are represented by boldface type, mononucleotide repeats (MNRs) are represented by underline, (-) represent missing nucleotides, and (*) represent nonconserved basepairs. Note that the isolates only vary by slight changes in the DNA nucleotide sequences. (b) Phylogenetic relationships and variation among 32 V. cholerae isolates.
So far, there have been over 200 serogroups of Vibrio cholerae identified based on variations in the O-antigen structure (Safa et. al., 2009). Based on phenotypic and genotypic differences, two major biotypes have been classified: Classical and El Tor. Both groups differ markedly in their pathogenic potentials, survivability, and infection patterns in human hosts (Safa et. al., 2009). However, in the last couple decades, several new isolates of Vibrio cholerae have emerged - the atypical El Tor strains (Figure 7a). These strains have traits of both the classical and the El Tor biotypes and may have evolved from El Tor variants through horizontal gene transfer mechanisms and genetic recombination events (Figure 7b). These atypical El Tor strains seem to be better adapted for dissemination, forcing the need for more measures to contain the toxin.
(a) 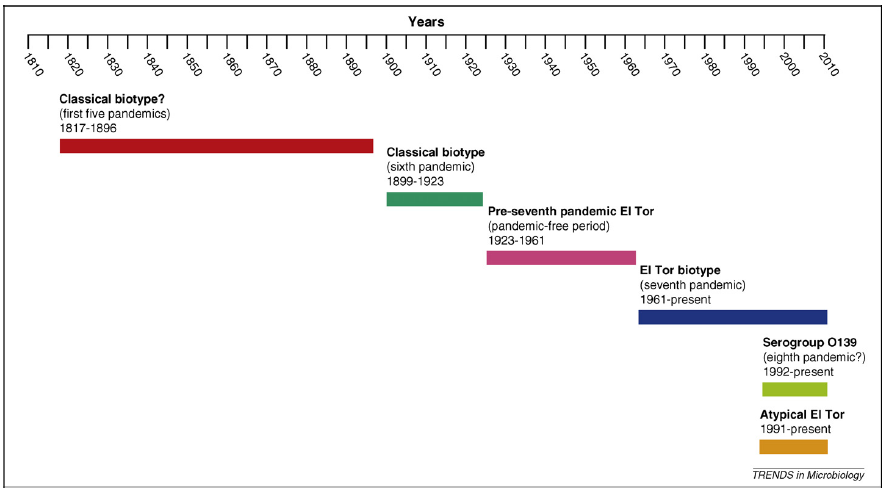 (b)
(b) 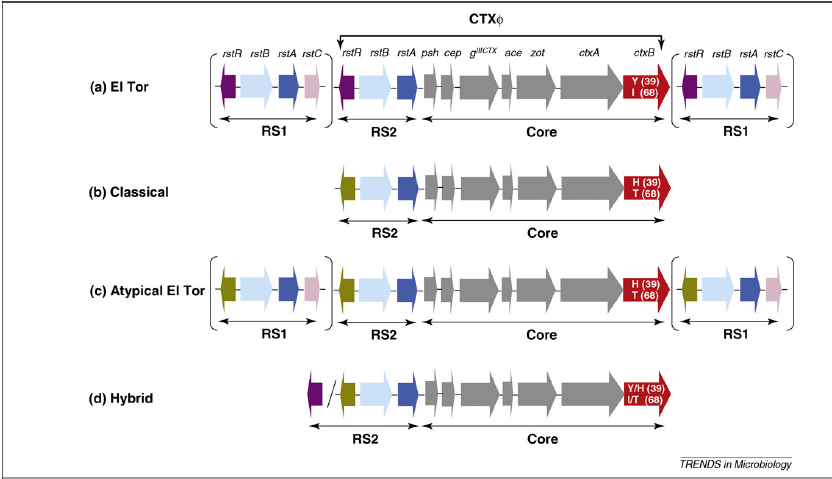
Obtained from Safa et. al., 2009. Permission pending
Figure 7. (a) Evolutionary events in cholera epidemiology since 1817. The atypical El Tor strains are characterized by a combination of "Classical" and 'El Tor' traits. (b) Structure of CTX and RS1 element in differentVibrio cholerae variants. The RS2 region encodes proteins for replication, integration, and regulation of site-specific recombination on the V. cholerae chromosome. The RS1 region is similar to RS2 but has an additional gene for antirepressor protein, which promotes the creation of infectious particles (Safa et. al., 2009). Note that the 'Classical' strain lacks the RS1 element, while the 'El Tor' and 'Atypical El Tor' strains contain the RS1 elements.
[1] Dallas W and Falkow S. 1980. Amino acid sequence homology between cholera toxin and Escherichia coli heat-labile toxin. Nature 288: 499-501.
[2] Danin-Poleg Y, Cohen L, Gancz H, Broza Y, Goldshmidt H, Malul E, Valinsky L, Lerner L, Broza M and Kashi Y. 2007. Vibrio cholerae strain typing and phylogeny study based on simple sequence repeats. Journal of Clinical Microbiology 45: 736-746.
[3] Koonin E. 2005. Orthologs, paralogs, and evolutionary genomics. Annual Review of Genetics 39: 309-338.
[4] Mekalanos J, Swartz D and Pearson G. 1983. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature 306: 551-557.
[5] Paradis S, Boissinot M, Paquette N, Belanger S, Martel E, Boudreau D, Picard F, Ouellette M, Roy P and Bergeron M. 2005. Phylogeny of the Enterobacteriaceae based on genes encoding elongation factor Tu and F-ATPase beta-subunit. International Journal of Systematic and Evolutionary Microbiology 55: 2013-2025.
[6] Peterson W and Ochoa L. 1989. Role of prostaglandins and cAMP in the secretory effects of cholera toxin. Science 245: 857-859.
[7] Safa A, Nair G and Kong R. 2009. Evolution of new variants of Vibrio cholerae O1. Cell: Trends in Microbiology 18: 46-54.
[8] Stine O, Sozhamannan S, Gou Q, Zheng S, Morris J and Johnson J. 2000. Phylogeny of Vibrio cholerae based on recA sequence. Infection and Immunity 68: 7180-7185.
[9] Yamamoto T, Nakazawa T, Miyata T, Kaji A and Yokota T. 1984. Evolution and structure of two ADP-ribosylation enterotoxins, Escherichia coli heat-labile toxin and cholera toxin. FEBS Letters 169: 241-246.
Please direct questions or comments to Teresa Wang