*This website was produced as an assignment for an undergraduate course at Davidson College
My Favorite Protein: α-amylase
What is α-amylase?
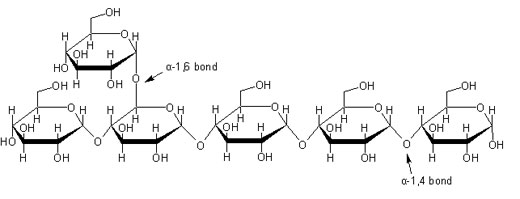
α-Amylase catalyzes the hydrolysis of α-1,4 glucan linkages in starch, one of the most abundant molecules on Earth essential to plant and animal energy storage (Brayer et al., 1995).

Figure 1. Representative starch structure indicating the α-1,4 linkage where α-amylase will target. Image from scienceinschool.org. Permission pending.
There are two kinds of α-amylases in humans, salivary and pancreatic amylase, which are isoenzymes. All mammalian species produce amylase in the pancrease, but the only mammals that produce salivary amylase are primates, rodents, and lagomorphs. Salivary and pancreatic amylases are encoded by distinct but closely related genes, AMY1A, AMY1B, AMY1C for salivary amylase and AMY2A and AMY2B for pancreatic amylase (Ting et al., 1992). In humans, α-amylase is encoded by a multigene family located on chromosome 1 (Gumucio et al., 1988). The regulation of these genes is such that different α-amylases are expressed in certain tissues in salivary glands and the pancrease. The pancreatic and salivary α-amylases are very similar with about a 97% homology overall (Brayer et al., 1995).
Structure of α-amylase:
The structure of human salivary α-amylase is found to have 496 amino-acid residues, one calcium ion, one chloride ion and 170 water molecules. Salivary amylase folds into a multidomain structure consisting of three domains, A, B, and C. Domain A has a (β/α) barrel structure, Domain B has no definite topology and Domain C has a Greek-key barrel structure seen in Figure 2. In salivary amylase, the calcium ion is bound to Asn100, Arg158, Asp167, His201 and three water molecules. The chloride ion is bound to Arg195, Asn298 and Arg337 and one water molecule, shown as a black ball in the middle of the molecule in Figure 2 (Ramasubbu et al., 1996).
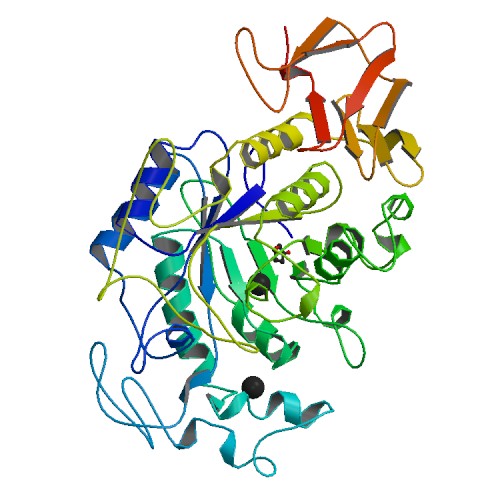
Figure 2. Crystal structure of human salivary α-amylase. Photo courtesy of PDB.
Similarly, human pancreatic α-amylase is found to be composed of three structural domains. As in salivary amylase, the largest domain is Domain A, which consists of an eight-stranded parallel β-barrel surrounded by a cylinder of α-helical segments. Domain A holds three putative active site residues Asp 197, Glu 233, and Asp 300. Bound near the active site is the chloride ion. Domain B is the smallest domain and primarily forms a β-structure. Domain B contains the calcium ion ligand. Domain C folds into an antiparallel β-barrel type structure and is much more loosely associated with Domain A than is Domain B. It is Domain C that is found to be the most variable between α-amylases of different origins, which can be seen when comparing the bottom portion of Figure 2 to the bottom portion of Figure 3. The protein groups making ligand interactions to calcium include Asn 100, Arg 158, Asp 167, and His 201, identical to salivary amylase. The protein groups making ligand interactions to chlorine include Arg 195, Asn 298, and Arg 337 (Bayer et al., 1995). These protein groups forming a ligand interaction with chlorine show significant difference than those from salivary amylase, indicating a possible change in activation site structure between the two proteins.
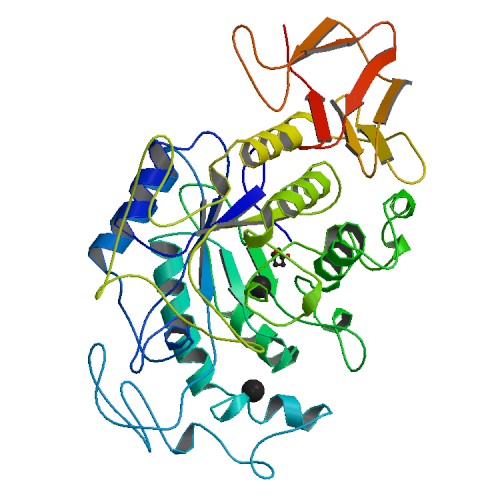
Figure 3. Crystal structure of human pancreatic α-amylase. Photo courtesy of PDB.
The salivary and pancreatic α-amylases are highly homologous in terms of primary sequence, but do exhibit somewhat different cleavage patterns. When comparing their amino acid sequences, Bayer and his colleagues found there are 15 amino acid differences between the two sequences. Overall the net effect of these substitutions is to add three more polar groups and three additional charged side chains to the salivary amylase. It was found that the largest amino acid change occurs in the segment of polypeptide chain that connects Domains A and C (Bayer et al., 1995).
Function of α-amylase:
Salivary and pancreatic α-amylases have similar functions. Both enzymes work in a slightly acidic pH and can digest starch quickly because they can bind anywhere on the substrate. The digestion of starch occurs in several stages in humans. Initially salivary amylase provides partial digestion, which breaks down polymeric starch into shorter oligomers including maltose and dextrin. When the partially digested starch reaches the gut, it is then extensively hydrolyzed into smaller oligosaccharides, like glucose, by the pancreatic amylase synthesized in the pancreas and excreted into the lumen (Bayer et al. 1995). The slight difference in function can possibly be attributed to the difference in active site and the slight difference in Domain C. The hydrolysis of starch by α-amylase is shown in Figure 4. Calcium and chloride are essential for both amylases in order to enhance stability of the molecule. Chloride, in particular appears to be vital for amylase to function because it is located near the active site of the enzyme, as mentioned above. Salivary amylase also has been found to have a special binding site allowing the molecule to bind to enamel (Ramasubbu et al., 1996).
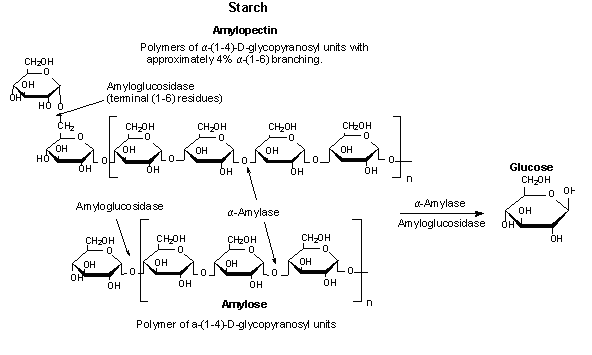
Figure 4. The hydrolysis of starch to glucose catalyzed by α-amylase. Image courtesy of Sigma-Aldrich. Permission pending.
References:
Brayer GD, Yaoguang L, Withers SG. The structure of human pancreatic α-amylase at 1.8 A resolution and comparisons with related enzymes. Protein Science 1995; 4: 1730-1742.
Gumucio DL, Wiebauer K, Caldwell RM, Samuelson LC, Meisler MH. Concerted evolution of human amylase genes. Mol Cell Biol 1998; 8: 1197-1205.
Ramasubbu N, Paloth V, Luo Y, Brayer GD, Levine MJ. Structure of human salivary α-amylase at 1.6 A resolution: Implications for its role in the oral cavity. Acta Crystallagraphica 1996; D52: 435-446.
Ting CN, Rosenberg MP, Snow CM, Samuelson LC, Meisler MH. Endogenous retroviral sequences are required for tissue-specific expression of a human salivary amylase gene. Public Access Comput Syst Rev [serial online] 1992; 6: 1457-1465. Avail from: genesdev.cshlp.org. Accessed 2010 Feb 16.
Please direct questions or comments to Abbey Webb