QuikChange
SITE-DIRECTED
MUTAGENESIS
***NOTE:
This information is purely for educational purposes. If you have any questions
about the product or ordering the product please contact:

USE
This method
allows a scientist to make small mutations in a piece of DNA. There
are three types of mutations that can be made - substitution, deletion,
or addition of nucleotides. The alteration in nucleotide sequence
then in turn alters the amino acid of the protein that it encodes for.
The mutation may or may not affect the function of the protein.
METHOD
In this procedure one
starts with miniprep plasmid DNA or cesium-chloride-purified DNA.
This isolates the desired DNA from others that may be in the "cocktail."
The starting DNA is double stranded with the gene of interest inserted
at some point. This gene of interest will be changed in some
fashion - by either inserting, deleting, or
substituting, a few base pairs. The starting DNA needs to be in the
form of a vector so that it may be transformed into E. coli cells in the
last step of the procedure.
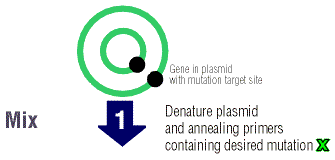
Next, single stranded chains of DNA (oligonucleotides) are
made complimentary to the opposite strands of the vector. These DNA strands
contain the desired substitution, deletion, or insertion. The oligonucleotides
are called primers because they will anneal to the plasmid after the two
strands have been denatured, or separated from each other. The primers
allow the Pfu Turbo polymerase to elongate the two DNA strands.
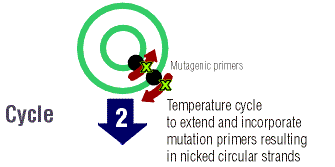 The parent DNA is elongated many times by temperature
cycling. At a high temperature, the DNA is denatured so that there are
two single strands upon which primers can anneal and the PfuTurbo DNA polymerase
can elongate the entire strand of DNA. There are dNTPs in the "cocktail"
that the polymerase uses to elongate the DNA strand. At the ends of each
DNA strand that is formed there are "staggered nicks" where the mutation
lies.
The parent DNA is elongated many times by temperature
cycling. At a high temperature, the DNA is denatured so that there are
two single strands upon which primers can anneal and the PfuTurbo DNA polymerase
can elongate the entire strand of DNA. There are dNTPs in the "cocktail"
that the polymerase uses to elongate the DNA strand. At the ends of each
DNA strand that is formed there are "staggered nicks" where the mutation
lies.
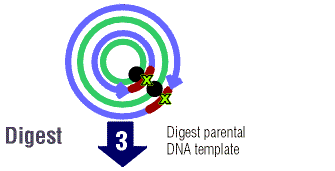 The new mutated DNA is treated and the parental DNA are treated
with Dpn I. Dpn I is an enzyme that digests methylated DNA. This
simply means that the DNA has methyl groups (CH3) attached to
it. Most bacterial strains do in fact methylate plasmid
DNA. It is very important the parent DNA be methylated so that the
enzyme can select for the new mutated DNA. (The new DNA will not
be mythylated.)
The new mutated DNA is treated and the parental DNA are treated
with Dpn I. Dpn I is an enzyme that digests methylated DNA. This
simply means that the DNA has methyl groups (CH3) attached to
it. Most bacterial strains do in fact methylate plasmid
DNA. It is very important the parent DNA be methylated so that the
enzyme can select for the new mutated DNA. (The new DNA will not
be mythylated.)
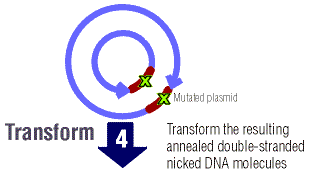 The nicked mutated DNA is then transformed into the E. coli cells.
Transformation is the uptake of foreign DNA by a bacteria cell
from the surrounding environment.1 The foreign DNA is
then incorporated into the genome of the bacteria and can be transcribed
and translated and altered.1
The nicked mutated DNA is then transformed into the E. coli cells.
Transformation is the uptake of foreign DNA by a bacteria cell
from the surrounding environment.1 The foreign DNA is
then incorporated into the genome of the bacteria and can be transcribed
and translated and altered.1
 After the E. coli cells are transformed, the XL1-Blue repairs the nicks
and connects the ends of the DNA stands.
After the E. coli cells are transformed, the XL1-Blue repairs the nicks
and connects the ends of the DNA stands.
WHY THIS METHOD?
ï
This method involves 4 steps that can be completed in one day. It
does not require a single stranded oligonucleotide as a template for elongation.
The double stranded DNA is simply denatured via high temperatures in order
to produce a template for the daughter cells. No unique restriction
sites are needed in the vector because the XL1-Blue E. coli repairs the
nicks in the daughter DNA and connects the two ends of the DNA.
ï
This method is very efficient and accurate. Pfu
DNA polymerase is used to elongate the daughter DNA strands. It provides
very accurate elongation with increased product. This method also provides
an alternative to PCR (polymerase chain reaction). Only the parent
strands are amplified in this procedure. The primers contain the
deletion, insertion, or substitution and are incorporated into the daughter
cell. In other words daughter DNA is never used as a template for "granddaughter"
DNA. This reduces the chances of unwanted mutations because the parent
DNA and primers always have the correct sequence. A mistake mutation
in a daughter cell will not be amplified. Additionally, more than
80% of the colonies are found to contain the selected DNA sequence when
plated out.
ï
This method works best with small numbers of base pair deletions, insertions,
or substitutions, usually between 1 and 12. There are optimum numbers
of base pair insertions, deletions, and substitutions and optimum numbers
of base pairs in the vectors used. For more specific information on these
numbers please click here
and scroll down to look at the chart for optimum number of base pairs in
certain situations.
Sources
1 Campbell, Neil A. Biology. 4th Ed. New York: The Benjamin/Cummings
Publishing Company, Inc., 1996. pg. 317 &338.
1999 February 11. PfuTurbo DNA Polymerase.<http://www.stratagene.com/pcr/pfuturbo.htm>
Accessed 1999
February 15.
1999 February 11. QuikChange Site-Directed Mutagenesis Kit.<http://www.stratagene.com/mutagenesis/quikchng.
htm>
Accessed 1999 February 15.
Trajano R.1999 February 11.Stratagene Online Home
Page.<http://www.stratagene.com>
Accessed 1999 February
15 .
Please send any questions or comments to : Sabrautigam@Davidson.edu
To go back to my homepage
click here





